1EXQ
 
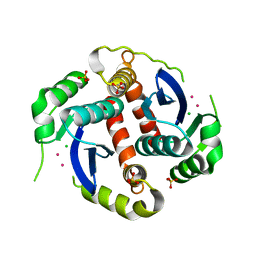 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1JXB
 
 | |
5DGW
 
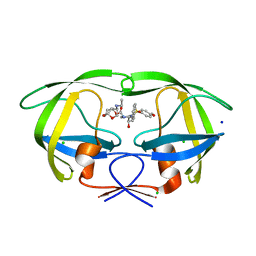 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | Descriptor: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
7DPQ
 
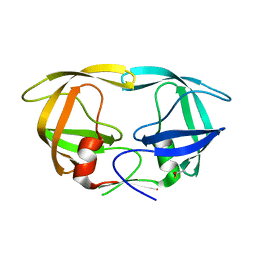 | | HIV-1 Protease D30N mutant | | Descriptor: | Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2020-12-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
J.Biomol.Struct.Dyn., 2021
|
|
2NPH
 
 | | Crystal structure of HIV1 protease in situ product complex | | Descriptor: | PROTEASE RETROPEPSIN, pentapeptide fragment, tetrapeptide fragment | | Authors: | Hosur, M.V, Das, A, Prashar, V. | | Deposit date: | 2006-10-27 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HIV-1 protease in situ product complex and observation of a low-barrier hydrogen bond between catalytic aspartates
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3KT2
 
 | | Crystal Structure of N88D mutant HIV-1 Protease | | Descriptor: | Protease | | Authors: | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
2WHH
 
 | | HIV-1 protease tethered dimer Q-product complex along with nucleophilic water molecule | | Descriptor: | GLUTAMIC ACID, PARA-NITROPHENYLALANINE, POL PROTEIN | | Authors: | Prashar, V, Bihani, S, Das, A, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-05-05 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Catalytic Water Co-Existing with a Product Peptide in the Active Site of HIV-1 Protease Revealed by X- Ray Structure Analysis.
Plos One, 4, 2009
|
|
3QIN
 
 | |
5XOS
 
 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
4U7Q
 
 | | Structure of wild-type HIV protease in complex with photosensitive inhibitor PDI-6 | | Descriptor: | N~2~-({[7-(diethylamino)-2-oxo-2H-chromen-4-yl]methoxy}carbonyl)-N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
3HYF
 
 | | Crystal structure of HIV-1 RNase H p15 with engineered E. coli loop and active site inhibitor | | Descriptor: | 2-(3,4-dichlorobenzyl)-5,6-dihydroxypyrimidine-4-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information.
J.Med.Chem., 52, 2009
|
|
7VSD
 
 | | E. coli Ribonuclease HI in complex with one Mg2+ (2) | | Descriptor: | MAGNESIUM ION, Ribonuclease HI, SULFATE ION | | Authors: | Liao, Z, Oyama, T. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pivotal role of a conserved histidine in Escherichia coli ribonuclease HI as proposed by X-ray crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5HRN
 
 | | HIV Integrase Catalytic Domain containing F185K mutation complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
7XJ5
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
6DJ5
 
 | | HIV-1 protease with mutation L76V in complex with GRL-0519 (tris-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
7VSA
 
 | | E. coli Ribonuclease HI in complex with two Mg2+ | | Descriptor: | GLYCEROL, MAGNESIUM ION, Ribonuclease HI, ... | | Authors: | Liao, Z, Oyama, T. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Pivotal role of a conserved histidine in Escherichia coli ribonuclease HI as proposed by X-ray crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5YRS
 
 | | X-ray Snapshot of HIV-1 Protease in Action: Observation of Tetrahedral Intermediate and Its SIHB with Catalytic Aspartate | | Descriptor: | PROTEASE, RT-RH oligopeprtide | | Authors: | Das, A, Mahale, S, Prashar, V, Bihani, S, Ferrer, J.-L, Hosur, M.V. | | Deposit date: | 2017-11-10 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
J. Am. Chem. Soc., 132, 2010
|
|
1JL2
 
 | |
6VQE
 
 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
3AA4
 
 | | A52V E.coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
4Q5M
 
 | | D30N tethered HIV-1 protease dimer/saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structural Basis of Why Nelfinavir-Resistant D30N Mutant of HIV-1 Protease Remains Susceptible to Saquinavir.
Chem.Biol.Drug Des., 86, 2015
|
|
1HWR
 
 | | MOLECULAR RECOGNITION OF CYCLIC UREA HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4-R-(4-ALPHA,6-BETA,7-BETA]-HEXAHYDRO-5,6-DI(HYDROXY)-1,3-DI(ALLYL)-4,7-BISPHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Chang, C.-H. | | Deposit date: | 1998-03-20 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular recognition of cyclic urea HIV-1 protease inhibitors.
J.Biol.Chem., 273, 1998
|
|
5KAO
 
 | | Crystal structure of wild type HIV-1 protease in complex with GRL-10413 | | Descriptor: | [(3~{a}~{S},4~{R},6~{a}~{R})-2,3,3~{a},4,5,6~{a}-hexahydrofuro[2,3-b]furan-4-yl] ~{N}-[(2~{S},3~{R})-1-(3-chloranyl-4-methoxy-phenyl)-4-[(4-methoxyphenyl)sulfonyl-(2-methylpropyl)amino]-3-oxidanyl-butan-2-yl]carbamate, protease | | Authors: | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-06-01 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.
Antimicrob.Agents Chemother., 60, 2016
|
|
1HVR
 
 | | RATIONAL DESIGN OF POTENT, BIOAVAILABLE, NONPEPTIDE CYCLIC UREAS AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-HEXAHYDRO-5,6-DIHYDROXY-1,3-BIS[2-NAPHTHYL-METHYL]-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPIN-2-ONE | | Authors: | Chang, C.-H. | | Deposit date: | 1994-02-14 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors.
Science, 263, 1994
|
|
1RBV
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
