1NBR
 
 | |
1NBS
 
 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | Descriptor: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | Authors: | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | Deposit date: | 2002-12-03 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
1NBU
 
 | | 7,8-Dihydroneopterin Aldolase Complexed with Product From Mycobacterium Tuberculosis | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, Probable dihydroneopterin aldolase | | Authors: | Goulding, C.W, Apostol, M.I, Sawaya, M.R, Phillips, M, Parseghian, A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-03 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme.
J.Mol.Biol., 349, 2005
|
|
1NBV
 
 | |
1NBW
 
 | | Glycerol dehydratase reactivase | | Descriptor: | CALCIUM ION, GLYCEROL DEHYDRATASE REACTIVASE ALPHA SUBUNIT, GLYCEROL DEHYDRATASE REACTIVASE BETA SUBUNIT | | Authors: | Liao, D.-I, Reiss, L, Turner Jr, I, Dotson, G. | | Deposit date: | 2002-12-04 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of glycerol dehydratase reactivase: A new type of molecular chaperone
Structure, 11, 2003
|
|
1NBX
 
 | | Streptavidin Mutant Y43A at 1.70A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NBY
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K96A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NBZ
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K97A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NC0
 
 | |
1NC1
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with 5'-methylthiotubercidin (MTH) | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1NC2
 
 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA | | Descriptor: | (S)-2-(4-(2-(2-HYDROXYETHYLTHIO)-ACETAMIDO)-BENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1NC4
 
 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Gd-DOTA | | Descriptor: | (S)-2-(4-NITROBENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
1NC5
 
 | | Structure of Protein of Unknown Function of YteR from Bacillus Subtilis | | Descriptor: | hypothetical protein yTER | | Authors: | Zhang, R, Lozondra, L, Korolev, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase.
Proteins, 60, 2005
|
|
1NC6
 
 | | Potent, small molecule inhibitors of human mast cell tryptase. Anti-asthmatic action of a dipeptide-based transition state analogue containing benzothiazole ketone | | Descriptor: | (2S,4R)-1-ACETYL-N-[(1S)-4-[(AMINOIMINOMETHYL)AMINO]-1-(2-BENZOTHIAZOLYLCARBONYL)BUTYL]-4-HYDROXY-2-PYRROLIDINECARBOXAMIDE, CALCIUM ION, SULFATE ION, ... | | Authors: | Costanzo, M.J, Yabut, S.C, Almond Jr, H.R, Andrade-Gordon, P, Corcoran, T.W, de Garavilla, L, Kauffman, J.A, Abraham, W.M, Recacha, R, Chattopadhyay, D, Maryanoff, B.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, Small-Molecule Inhibitors of Human Mast Cell Tryptase. Antiasthmatic Action of a Dipeptide-Based Transition-State Analogue Containing a Benzothiazole Ketone.
J.Med.Chem., 46, 2003
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
1NC8
 
 | | HIGH-RESOLUTION SOLUTION NMR STRUCTURE OF THE MINIMAL ACTIVE DOMAIN OF THE HUMAN IMMUNODEFICIENCY VIRUS TYPE-2 NUCLEOCAPSID PROTEIN, 15 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Kodera, Y, Sato, K, Tsukahara, T, Komatsu, H, Maeda, T, Kohno, T. | | Deposit date: | 1998-05-14 | | Release date: | 1999-05-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein.
Biochemistry, 37, 1998
|
|
1NC9
 
 | | STREPTAVIDIN MUTANT Y43A WITH IMINOBIOTIN AT 1.8A RESOLUTION | | Descriptor: | 2-IMINOBIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-05 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NCA
 
 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NCB
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1NCC
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1NCD
 
 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NCE
 
 | | Crystal structure of a ternary complex of E. coli thymidylate synthase D169C with dUMP and the antifolate CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2002-12-05 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The only active mutant of thymidylate synthase D169, a residue far from the
site of methyl transfer, demonstrates the exquisite nature of enzyme
specificity.
Protein Eng., 16, 2003
|
|
1NCF
 
 | |
1NCG
 
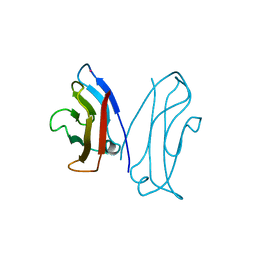 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
