2Y28
 
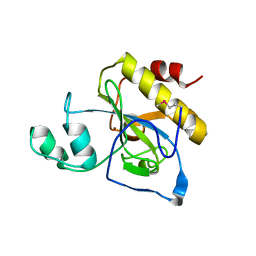 | | crystal structure of Se-Met AmpD derivative | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
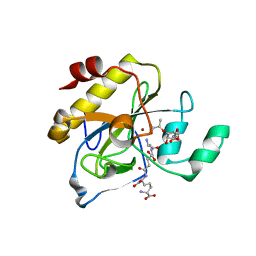 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
7Q4B
 
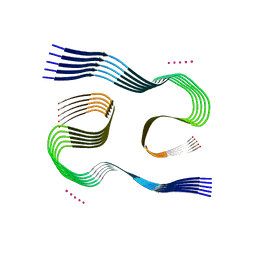 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7MOO
 
 | | L-type DNA containing 2'-fluoro-2'-deoxycytidine | | Descriptor: | DNA (5'-D(*(0DG)P*(OFC)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DG)P*(0DC))-3') | | Authors: | Dantsu, Y, Zhang, Y, Zhang, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis of 2'-Deoxy-2'-fluoro-L-cytidine and Fluorinated L-Nucleic Acids for Structural Studies
Chemistryselect, 6, 2021
|
|
8TF7
 
 | |
5TJB
 
 | | I-II linker of TRPML1 channel at pH 4.5 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7Q4M
 
 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
6J1U
 
 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
8FZC
 
 | |
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7QIG
 
 | | Infectious mouse-adapted RML scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Zhang, W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2021-12-14 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | 2.7 angstrom cryo-EM structure of ex vivo RML prion fibrils.
Nat Commun, 13, 2022
|
|
3EWT
 
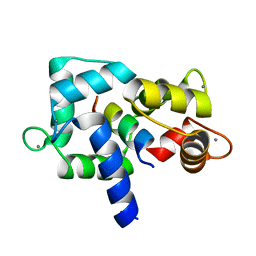 | | Crystal Structure of calmodulin complexed with a peptide | | Descriptor: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | Deposit date: | 2008-10-16 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7L97
 
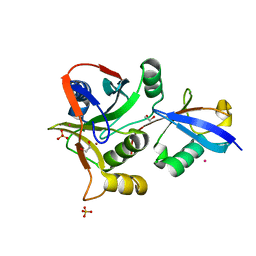 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
5WWO
 
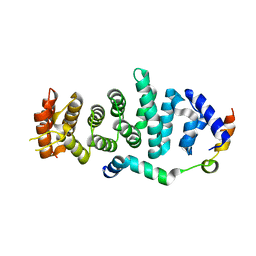 | | Crystal structure of Enp1 | | Descriptor: | Essential nuclear protein 1, Protein LTV1 | | Authors: | Ye, K, Zhang, W. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
3EWV
 
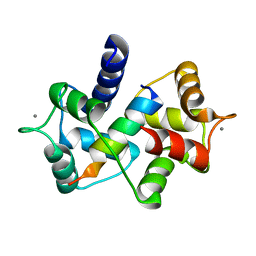 | | Crystal Structure of calmodulin complexed with a peptide | | Descriptor: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | Deposit date: | 2008-10-16 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8F24
 
 | | Mirror-image RNA octamer containing 2'-OMe-L-uridine | | Descriptor: | Mirror-image RNA 0G-XEC-0G-0U-0A-0C-0A-0C, SULFATE ION | | Authors: | Dantsu, Y, Zhang, W. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
6J36
 
 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
9J56
 
 | | Functional Investigation of the SAM-Dependent Methyltransferases Rdmb in Anthracycline Biosynthesis | | Descriptor: | (7~{S},9~{S})-7-[(2~{R},4~{S},5~{S},6~{S})-4-azanyl-6-methyl-5-oxidanyl-oxan-2-yl]oxy-9-ethyl-4-methoxy-6,9,11-tris(oxidanyl)-8,10-dihydro-7~{H}-tetracene-5,12-dione, Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Q.Y, Sang, M.L, Zhang, W. | | Deposit date: | 2024-08-11 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional investigation of the SAM-dependent methyltransferase RdmB in anthracycline biosynthesis.
Synth Syst Biotechnol, 10, 2025
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4CA9
 
 | | Structure of the Nucleoplasmin-like N-terminal domain of Drosophila FKBP39 | | Descriptor: | 39 KDA FK506-BINDING NUCLEAR PROTEIN | | Authors: | Artero, J, Forsyth, T, Callow, P, Watson, A.A, Zhang, W, Laue, E.D, Edlich-Muth, C, Przewloka, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
J.Mol.Biol., 427, 2015
|
|
4BOL
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4BPA
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
8CN0
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNB
 
 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
