5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Rang, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
5WVN
 
 | | Crystal structure of MBS-BaeS fusion protein | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Ran, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
6A2W
 
 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
8K8T
 
 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
3UTD
 
 | | Ec_IspH in complex with 4-oxopentyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-oxopentyl trihydrogen diphosphate, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UV7
 
 | | Ec_IspH in complex with buta-2,3-dienyl diphosphate (1300) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, buta-2,3-dien-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
2XUV
 
 | | The structure of HdeB | | Descriptor: | HDEB, SULFATE ION | | Authors: | Naismith, J.H, Wang, W. | | Deposit date: | 2010-10-21 | | Release date: | 2011-08-24 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Salt Bridges Regulate Both Dimer Formation and Monomeric Flexibility in Hdeb and May Have a Role in Periplasmic Chaperone Function.
J.Mol.Biol., 415, 2012
|
|
4FK3
 
 | | B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 | | Descriptor: | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Wang, W, Zhang, K.Y.J. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7MDP
 
 | | KRas G12C in complex with G-2897 | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 4-(trifluoromethyl)-1,3-benzothiazol-2-amine, ... | | Authors: | Oh, A, Frank, Y, Wang, W. | | Deposit date: | 2021-04-05 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
5VRQ
 
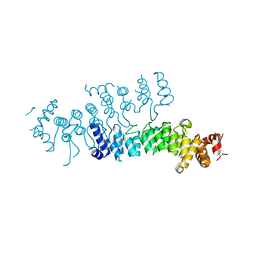 | | Crystal structure of Legionella pneumophila effector AnkC | | Descriptor: | Ankyrin repeat-containing protein | | Authors: | Kozlov, G, Wong, K, Wang, W, Skubak, P, Munoz-Escobar, J, Liu, Y, Pannu, N.S, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Ankyrin repeats as a dimerization module.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5VZY
 
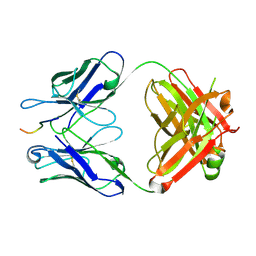 | | Crystal structure of crenezumab Fab in complex with Abeta | | Descriptor: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5VZX
 
 | | Crystal structure of crenezumab Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Crenezumab Fab heavy chain, ... | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5WBS
 
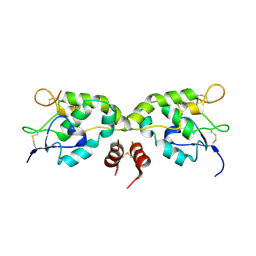 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
4O10
 
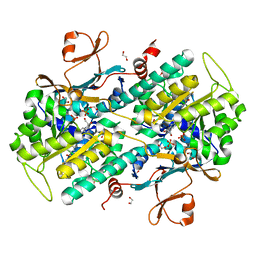 | | Structural and Biochemical Analyses of the Catalysis and Potency Impact of Inhibitor Phosphoribosylation by Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}indolizine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
4O18
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O0V
 
 | |
4O17
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1A
 
 | | The crystal structure of the mutant NAMPT G217R | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O28
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-17 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1B
 
 | | The crystal structure of a mutant NAMPT (G217R) in complex with an inhibitor APO866 | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O12
 
 | |
4O0X
 
 | | Back pocket flexibility provides group-II PAK selectivity for type 1 kinase inhibitors | | Descriptor: | 1-{[1-(4-amino-1,3,5-triazin-2-yl)-2-methyl-1H-benzimidazol-6-yl]ethynyl}cyclohexanol, Serine/threonine-protein kinase PAK 4 | | Authors: | Rouge, L, Tam, C, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-02-12 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Back Pocket Flexibility Provides Group II p21-Activated Kinase (PAK) Selectivity for Type I 1/2 Kinase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
