7CN4
 
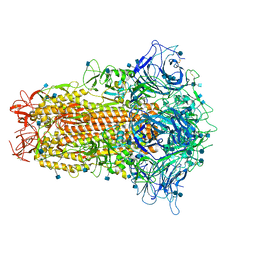 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | 登録日 | 2020-07-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
6KEA
 
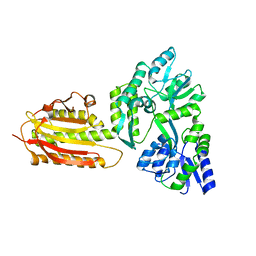 | | crystal structure of MBP-tagged REV7-IpaB complex | | 分子名称: | Maltose-binding periplasmic protein,LINKER,hREV7,LINKER,Invasin IpaB,hREV3 | | 著者 | Wang, X, Pernicone, N, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | 登録日 | 2019-07-04 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
7DH6
 
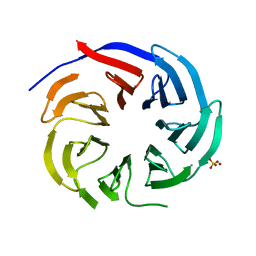 | | Crystal structure of PLRG1 | | 分子名称: | CALCIUM ION, NICKEL (II) ION, Pleiotropic regulator 1, ... | | 著者 | Wang, X, Xu, C. | | 登録日 | 2020-11-13 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.584 Å) | | 主引用文献 | Crystal structure of the WD40 domain of human PLRG1.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7CYH
 
 | |
6L77
 
 | |
7BIK
 
 | | Crystal structure of YTHDF2 in complex with m6Am | | 分子名称: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-4-methoxy-5-[6-(methylamino)purin-9-yl]oxolan-3-ol, GLYCEROL, SULFATE ION, ... | | 著者 | Wang, X, Caflisch, A. | | 登録日 | 2021-01-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of YTHDF2 in complex with m6Am
To Be Published
|
|
6M16
 
 | | Cryo-EM structures of SADS-CoV spike glycoproteins | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Wang, X, Yu, J, Qiao, S, Guo, R. | | 登録日 | 2020-02-24 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
6M15
 
 | | Cryo-EM structures of HKU2 spike glycoproteins | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Wang, X, Yu, J, Qiao, S, Guo, R. | | 登録日 | 2020-02-24 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.38 Å) | | 主引用文献 | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
7VHL
 
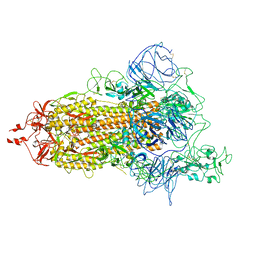 | |
7VHK
 
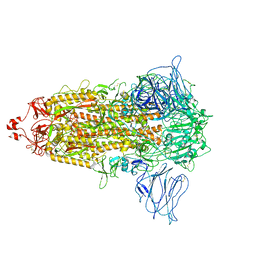 | |
7VHJ
 
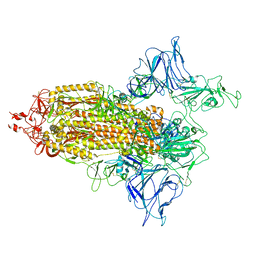 | |
7VHN
 
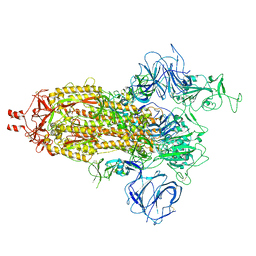 | |
7X39
 
 | | Structure of CIZ1 bound ERH | | 分子名称: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | 著者 | Wang, X, Xu, C. | | 登録日 | 2022-02-28 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
1QFD
 
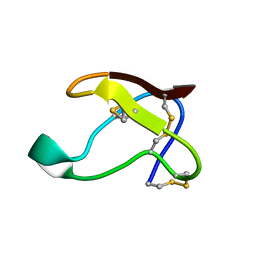 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | 分子名称: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | 著者 | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | 登録日 | 1999-04-08 | | 公開日 | 1999-07-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
3J1F
 
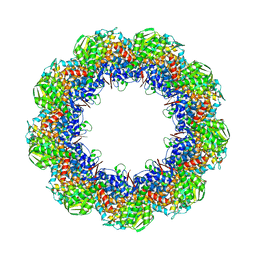 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in ATP-binding state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin beta subunit, MAGNESIUM ION | | 著者 | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | 登録日 | 2012-02-06 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
2ACV
 
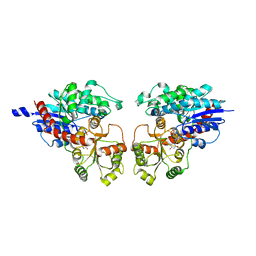 | | Crystal Structure of Medicago truncatula UGT71G1 | | 分子名称: | URIDINE-5'-DIPHOSPHATE, triterpene UDP-glucosyl transferase UGT71G1 | | 著者 | Shao, H, He, X, Achnine, L, Blount, J.W, Dixon, R.A, Wang, X. | | 登録日 | 2005-07-19 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago truncatula.
Plant Cell, 17, 2005
|
|
9FT1
 
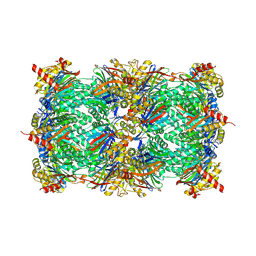 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FST
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor LU-001i | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-21 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | 分子名称: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | 分子名称: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-22 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
6PET
 
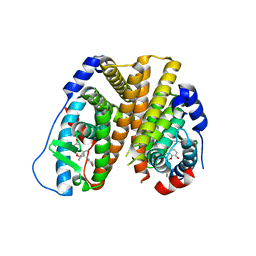 | | Crystal structure of 8-hydroxychromene compound 30 bound to estrogen receptor alpha | | 分子名称: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-8-ol, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, CHLORIDE ION, ... | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Wang, X, Zbieg, J, Labadie, S.S, Zhang, B, Li, J, Liang, W. | | 登録日 | 2019-06-20 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.203 Å) | | 主引用文献 | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6PFM
 
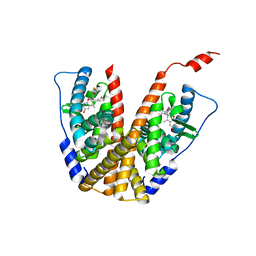 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | 分子名称: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | 登録日 | 2019-06-21 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1ZYR
 
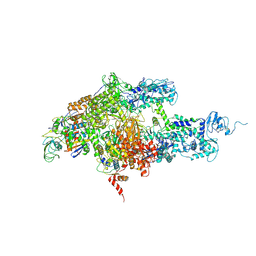 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | 分子名称: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | 著者 | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | 登録日 | 2005-06-10 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
4X0V
 
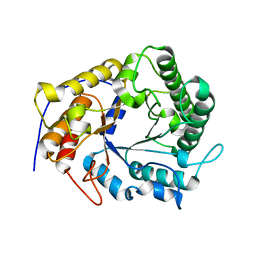 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | 分子名称: | Beta-1,3-1,4-glucanase | | 著者 | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | 登録日 | 2014-11-24 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.798 Å) | | 主引用文献 | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
4WQN
 
 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | 著者 | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | 登録日 | 2014-10-22 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.121 Å) | | 主引用文献 | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
