8Q4L
 
 | | GBP1 bound by 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, Guanylate-binding protein 1 | | Authors: | Pfleiderer, M.M, Liu, X, Fisch, D, Anastasakou, E, Frickel, E.M, Galej, W.P. | | Deposit date: | 2023-08-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (5.12 Å) | | Cite: | PIM1 controls GBP1 activity to limit self-damage and to guard against pathogen infection.
Science, 382, 2023
|
|
6EG8
 
 | | Structure of the GDP-bound Gs heterotrimer | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hilger, D, Liu, X, Aschauer, P, Kobilka, B.K. | | Deposit date: | 2018-08-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
1FAW
 
 | | GRAYLAG GOOSE HEMOGLOBIN (OXY FORM) | | Descriptor: | HEMOGLOBIN (ALPHA SUBUNIT), HEMOGLOBIN (BETA SUBUNIT), OXYGEN MOLECULE, ... | | Authors: | Liang, Y.-H, Liu, X.-Z, Liu, S.-H, Lu, G.-Y. | | Deposit date: | 2000-07-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The structure of greylag goose oxy haemoglobin: the roles of four mutations compared with bar-headed goose haemoglobin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5X62
 
 | |
5X4H
 
 | | The crystal structure of Pyrococcus furiosus RecJ (wild-type) | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4I
 
 | | Pyrococcus furiosus RecJ (D83A, Mn-soaking) | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
6KY5
 
 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
2AIU
 
 | | Crystal Structure of Mouse Testicular Cytochrome C at 1.6 Angstrom | | Descriptor: | Cytochrome c, testis-specific, PHOSPHATE ION, ... | | Authors: | Liu, Z, Ye, S, Lin, H, Rao, Z, Liu, X.J. | | Deposit date: | 2005-08-01 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Remarkably high activities of testicular cytochrome c in destroying reactive oxygen species and in triggering apoptosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6KIK
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | Descriptor: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
5BJS
 
 | | Apo ctPRC2 in an autoinhibited state | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein SUZ12, Polycomb Protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2016-10-22 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
6KY6
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | Descriptor: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KHP
 
 | | Crystal structure of Oryza sativa TDC with PLP and tryptamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, ACETATE ION, CALCIUM ION, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
6KIY
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KTR
 
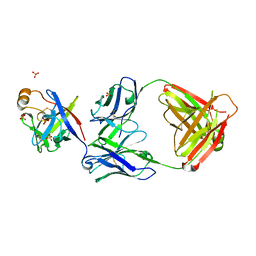 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
4X0V
 
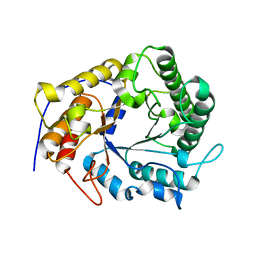 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | Descriptor: | Beta-1,3-1,4-glucanase | | Authors: | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
1VFQ
 
 | |
6K50
 
 | |
4RL2
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (1R)-2-({(R)-carboxy[(2R,5S)-4-carboxy-5-methyl-5,6-dihydro-2H-1,3-thiazin-2-yl]methyl}amino)-2-oxo-1-phenylethanaminium, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RL0
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
1DXA
 
 | | BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA (5'-D(*CP*TP*CP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*AP*CP*GP*AP*G)-3') | | Authors: | Yeh, H.J.C, Sayer, J.M, Liu, X, Altieri, A.S, Byrd, R.A, Lakshman, M.K, Yagi, H, Schurter, E.J, Gorenstein, D.G, Jerina, D.M. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10S adduct derived from trans addition of a deoxyadenosine N6-amino group to (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10- tetrahydrobenzo[a]pyrene: an unusual syn glycosidic torsion angle at the modified dA
Biochemistry, 34, 1995
|
|
4RM5
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
5F5O
 
 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
6K4V
 
 | | The solution structure of the smart chimeric peptide G6 | | Descriptor: | smart chimeric peptide G6 | | Authors: | Wang, J.H, Liu, X.H. | | Deposit date: | 2019-05-27 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli.
Commun Biol, 3, 2020
|
|
7YUY
 
 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
6K4W
 
 | | smart chimeric peptide SCP-A6 | | Descriptor: | SCP-A6 | | Authors: | Wang, J.H, Liu, X.H. | | Deposit date: | 2019-05-27 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli.
Commun Biol, 3, 2020
|
|
