8U3C
 
 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 2 (monomer) | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8FNE
 
 | | phiPA3 PhuN Tetramer, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
7L2O
 
 | |
7L2K
 
 | |
7L2U
 
 | | cryo-EM structure of DkTx-bound minimal TRPV1 in open state | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2H
 
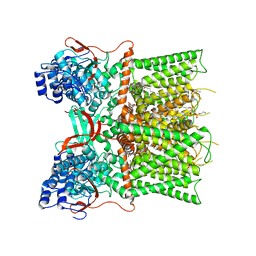 | | Cryo-EM structure of unliganded full-length TRPV1 at neutral pH | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl tridecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2J
 
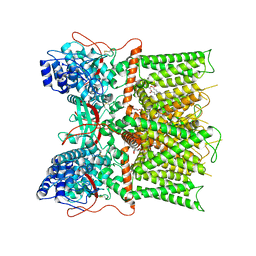 | | Cryo-EM structure of full-length TRPV1 at pH6c state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2R
 
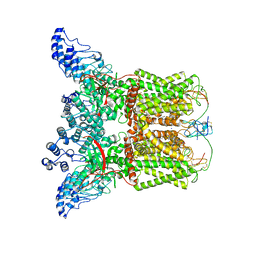 | | Cryo-EM structure of DkTx-bound minimal TRPV1 at the pre-open state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2M
 
 | | Cryo-EM structure of DkTx/RTX-bound full-length TRPV1 | | Descriptor: | SODIUM ION, Tau-theraphotoxin-Hs1a, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2I
 
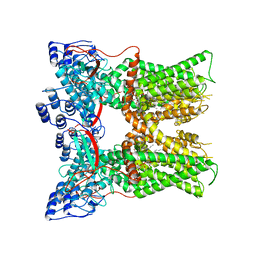 | | Cryo-EM structure of full-length TRPV1 at pH6a state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2W
 
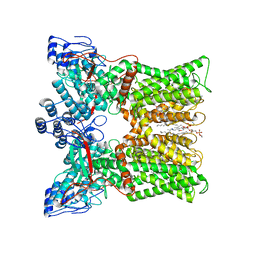 | | cryo-EM structure of RTX-bound minimal TRPV1 with NMDG at state a | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2V
 
 | | cryo-EM structure of RTX-bound minimal TRPV1 with NMDG at state b | | Descriptor: | 6-deoxy-6-(methylamino)-D-galactitol, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2S
 
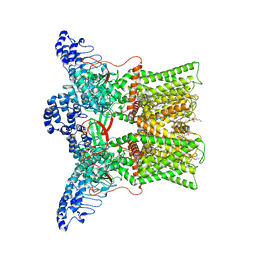 | | cryo-EM structure of DkTx-bound minimal TRPV1 at the pre-bound state | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2P
 
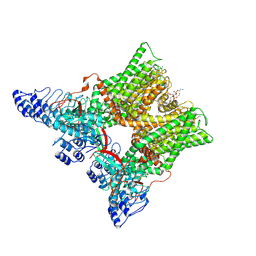 | | cryo-EM structure of unliganded minimal TRPV1 | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2N
 
 | |
7L2X
 
 | | cryo-EM structure of RTX-bound minimal TRPV1 with NMDG at state c | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, 1-deoxy-1-(methylamino)-D-allitol, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2L
 
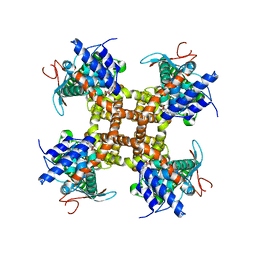 | |
7L2T
 
 | | cryo-EM structure of DkTx-bound minimal TRPV1 in partial open state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
6JHY
 
 | |
7T32
 
 | | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein | | Authors: | Zhang, K.H, Wu, H, Hoppe, N, Manglik, A, Cheng, Y.F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
7RMI
 
 | | SP6-11 biased agonist bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMG
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMH
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
6O6A
 
 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, ligand-free state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, SODIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
4HVY
 
 | | A thermostable variant of human NUDT18 NUDIX domain obtained by Hot Colony Filtration | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate-linked moiety X motif 18, ... | | Authors: | Asial, I, Cheng, Y.X, Engman, H, Wu, B, Dollhopf, M, Nordlund, P, Cornvik, T. | | Deposit date: | 2012-11-07 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell.
Nat Commun, 4, 2013
|
|
