6XDG
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
1C1J
 
 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
3OOP
 
 | | The structure of a protein with unknown function from Listeria innocua Clip11262 | | Descriptor: | Lin2960 protein | | Authors: | Fan, Y, Li, H, Zhou, Y, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a protein with unknown function from Listeria innocua Clip11262
To be Published
|
|
1KGS
 
 | |
2AY0
 
 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | Descriptor: | Bifunctional putA protein, CHLORIDE ION | | Authors: | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | Deposit date: | 2005-09-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
8UQQ
 
 | | Structure of mCLIFY: a circularly permuted yellow fluorescent protein | | Descriptor: | mCLIFY | | Authors: | Shweta, H, Gupta, K, Zhou, Y, Cui, X, Li, S, Lu, Z, Goldman, Y.E, Dantzig, J. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of mCLIFY: a circularly permuted yellow fluorescent protein
To Be Published
|
|
1JVM
 
 | |
8XEP
 
 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | Descriptor: | SULFATE ION, Type IV effector MavL, Ubiquitin | | Authors: | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
3W4R
 
 | | Crystal structure of an insect chitinase from the Asian corn borer, Ostrinia furnacalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Shen, X, Yang, Q. | | Deposit date: | 2013-01-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL0
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKZ
 
 | | Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL1
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3X21
 
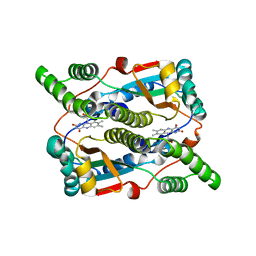 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
3X22
 
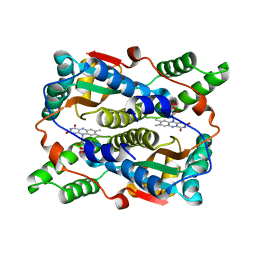 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
2DGN
 
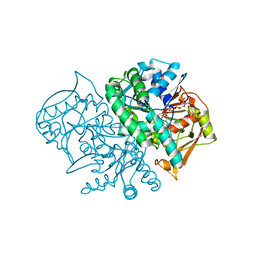 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
3VTR
 
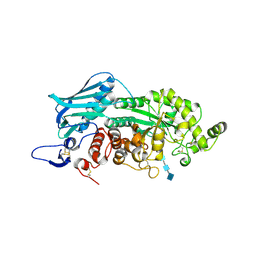 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | Deposit date: | 2012-06-02 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
2J6P
 
 | | STRUCTURE OF AS-SB REDUCTASE FROM LEISHMANIA MAJOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SB(V)-AS(V) REDUCTASE, ... | | Authors: | Bisacchi, D, Zhou, Y, Rosen, B.P, Mukhopadhyay, R, Bordo, D. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the As/Sb reductase LmACR2 from Leishmania major.
J. Mol. Biol., 386, 2009
|
|
2GIZ
 
 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
3WZF
 
 | | Crystal structure of human cytoplasmic aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Jiang, X, Chang, H, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2014-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Recombinant expression, purification and Preliminary crystallographic studies of human cytoplasmic aspartate aminotransferase
To be Published
|
|
6PDX
 
 | |
3WMC
 
 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(dimethylamino)-2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Beta-hexosaminidase | | Authors: | Chen, L, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
3WMB
 
 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 | | Descriptor: | 2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
5GQB
 
 | | Crystal structure of chitinase-h from O. furnacalis in complex with chitohepatose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure, Catalysis, and Inhibition of OfChi-h, the Lepidoptera-exclusive Insect Chitinase.
J. Biol. Chem., 292, 2017
|
|
5GPR
 
 | | Crystal structure of chitinase-h from Ostrinia furnacalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure, Catalysis, and Inhibition of OfChi-h, the Lepidoptera-exclusive Insect Chitinase.
J. Biol. Chem., 292, 2017
|
|
