7QN5
 
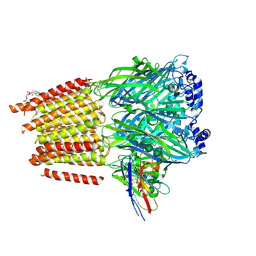 | | Cryo-EM structure of human full-length extrasynaptic alpha4beta3delta GABA(A)R in complex with nanobody Nb25 | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QN7
 
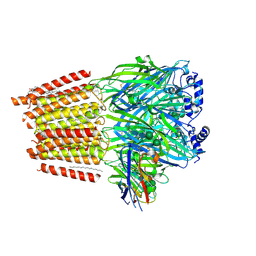 | | Cryo-EM structure of human full-length extrasynaptic alpha4beta3delta GABA(A)R in complex with GABA, histamine and nanobody Nb25 | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNE
 
 | | Cryo-EM structure of human full-length synaptic alpha1beta3gamma2 GABA(A)R in complex with Ro15-4513 and megabody Mb38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
8GS8
 
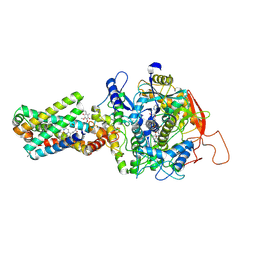 | | cryo-EM structure of the human respiratory complex II | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Du, Z, Zhou, X, Lai, Y, Xu, J, Zhang, Y, Zhou, S, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the human respiratory complex II.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4GAZ
 
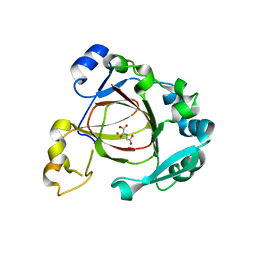 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | Descriptor: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
6VN7
 
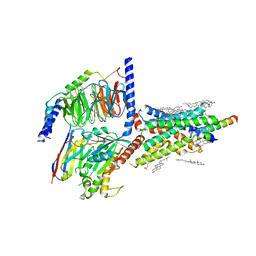 | | Cryo-EM structure of an activated VIP1 receptor-G protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Duan, J, Shen, D.-D, Zhou, X.E, Liu, Q.-F, Zhuang, Y.-W, Zhang, H.-B, Xu, P.-Y, Ma, S.-S, He, X.-H, Melcher, K, Zhang, Y, Xu, H.E, Yi, J. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy.
Nat Commun, 11, 2020
|
|
8F7S
 
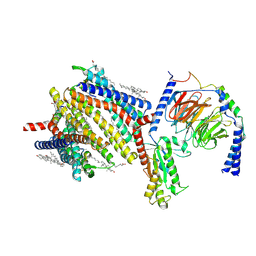 | | Gi bound delta-opioid receptor in complex with deltorphin | | Descriptor: | CHOLESTEROL, Delta-type opioid receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, DiBerto, J.F, Zhou, X.E, Schmitz, G.P, Yuan, Q, Jain, M.K, Liu, W, Melcher, K, Jiang, Y, Roth, B.L, Xu, H.E. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the entire human opioid receptor family.
Cell, 186, 2023
|
|
6OMM
 
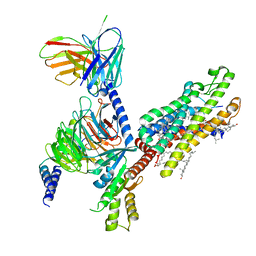 | | Cryo-EM structure of formyl peptide receptor 2/lipoxin A4 receptor in complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Liu, H, de Waal, P.W, Zhou, X.E, Wang, L, Meng, X, Zhao, G, Kang, Y, Melcher, K, Xu, H.E, Zhang, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of formylpeptide receptor 2-Gicomplex reveals insights into ligand recognition and signaling.
Nat Commun, 11, 2020
|
|
4LA1
 
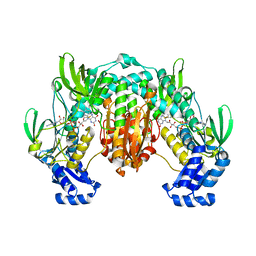 | | Crystal structure of SjTGR (thioredoxin glutathione reductase from Schistosoma japonicumi)complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Peng, Y, Wu, Q, Huang, F, Chen, J, Li, X, Zhou, X, Fan, X. | | Deposit date: | 2013-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of SjTGR complex with FAD
To be Published
|
|
7Y9B
 
 | | Crystal structure of the membrane (M) protein of a SARS-COV-2-related coronavirus | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, Membrane protein | | Authors: | Wang, X, Sun, Z, Zhou, X. | | Deposit date: | 2022-06-24 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Crystal structure of the membrane (M) protein from a bat betacoronavirus.
Pnas Nexus, 2, 2023
|
|
2HPL
 
 | | Crystal structure of the mouse p97/PNGase complex | | Descriptor: | C-terminal of mouse p97/VCP, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HPJ
 
 | | Crystal structure of the PUB domain of mouse PNGase | | Descriptor: | GLYCEROL, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7Y96
 
 | |
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
2F4O
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, PHQ-VAL-ALA-ASP-CF0, XP-C repair complementing complex 58 kDa protein, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
7YGQ
 
 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
1PN9
 
 | | Crystal structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae | | Descriptor: | Glutathione S-transferase 1-6, S-HEXYLGLUTATHIONE | | Authors: | Chen, L, Hall, P.R, Zhou, X.E, Ranson, H, Hemingway, J, Meehan, E.J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO7
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TDH
 
 | | The crystal structure of the dominant negative mutant G protein alpha(i)-1-beta-1-gamma-2 G203A/A326S | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, P, Jia, M.-Z, Zhou, X.E, de Waal, P.W, Dickson, B.M, Liu, B, Hou, L, Yin, Y.-T, Kang, Y.-Y, Shi, Y, Melcher, K, Xu, H.E, Jiang, Y. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of the dominant negative phenotype of the G alpha i1 beta 1 gamma 2 G203A/A326S heterotrimer
Acta Pharmacol.Sin., 37, 2016
|
|
5TVB
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TWO
 
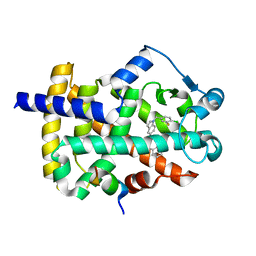 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selectively PPAR gamma-modulating ligand VSP-51 | | Descriptor: | N-benzyl-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-5-carboxamide, PRO-SER-LEU-LEU-LYS-LYS-LEU-LEU-LEU-ALA-PRO, Peroxisome proliferator-activated receptor gamma | | Authors: | Yi, W, Shi, J, Zhao, G, Zhou, X.E, Suino-Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-14 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Identification of a novel selective PPAR gamma ligand with a unique binding mode and improved therapeutic profile in vitro.
Sci Rep, 7, 2017
|
|
6OE8
 
 | | The crystal structure of hyper-thermostable AgUricase mutant K12C/E286C | | Descriptor: | MALONATE ION, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Shi, Y, Wang, T, Zhou, X.E, Liu, Q, Jiang, Y, Xu, H.E. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-based design of a hyperthermostable AgUricase for hyperuricemia and gout therapy.
Acta Pharmacol.Sin., 40, 2019
|
|
5H3O
 
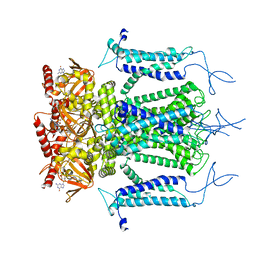 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
