3HJY
 
 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*GP*UP*GP*CP*GP*GP*UP*UP*U)-3', 5'-R(*GP*GP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*CP*CP*GP*CP*GP*GP*CP*GP*C)-3', RNA (25-MER), ... | | Authors: | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HJW
 
 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(FHU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, POTASSIUM ION, ... | | Authors: | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
5H2T
 
 | |
3R4U
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1 | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
6VPY
 
 | | I33M (I3.2 mutant from CH103 Lineage) | | Descriptor: | CHLORIDE ION, GLYCEROL, I33M heavy chain, ... | | Authors: | Fera, D, Zhou, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Effects of Framework Mutations at the Variable Domain Interface on Antibody Affinity Maturation in an HIV-1 Broadly Neutralizing Antibody Lineage.
Front Immunol, 11, 2020
|
|
3R4S
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1, N-acetyl-alpha-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
5H5X
 
 | |
1CW3
 
 | | HUMAN MITOCHONDRIAL ALDEHYDE DEHYDROGENASE COMPLEXED WITH NAD+ AND MN2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, MITOCHONDRIAL ALDEHYDE DEHYDROGENASE, ... | | Authors: | Ni, L, Zhou, J, Hurley, T.D, Weiner, H. | | Deposit date: | 1999-08-25 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms.
Protein Sci., 8, 1999
|
|
6M61
 
 | | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with inhibitor heptelidic acid | | Descriptor: | (5aS,6R,9S,9aS)-9-methyl-9-oxidanyl-1-oxidanylidene-6-propan-2-yl-3,5a,6,7,8,9a-hexahydro-2-benzoxepine-4-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82449543 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6M10
 
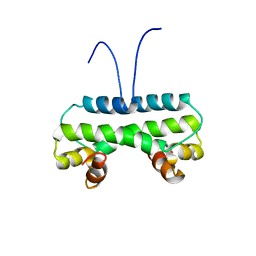 | | Crystal structure of PA4853 (Fis) from Pseudomonas aeruginosa | | Descriptor: | Putative Fis-like DNA-binding protein | | Authors: | Zhang, H, Gao, Z, Zhou, J, Dong, Y. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of the nucleoid-associated protein Fis (PA4853) from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6M5X
 
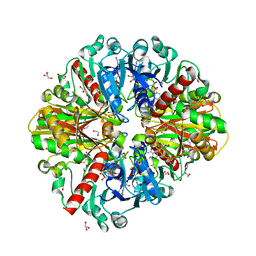 | | A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05990934 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6N1D
 
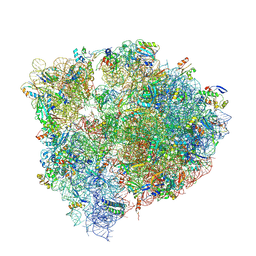 | | X-ray Crystal complex showing Spontaneous Ribosomal Translocation of mRNA and tRNAs into a Chimeric Hybrid State | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noller, H.F, Donohue, J.P, Lancaster, L, Zhou, J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2HZ6
 
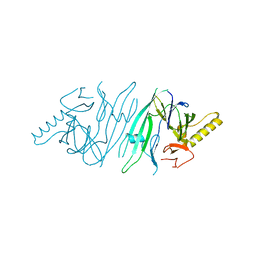 | | The crystal structure of human IRE1-alpha luminal domain | | Descriptor: | Endoplasmic reticulum to nucleus signalling 1 isoform 1 variant | | Authors: | Kaufman, R.J, Xu, Z, Zhou, J. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6JJ7
 
 | | Crystal structure of OsHXK6-Glc complex | | Descriptor: | Rice hexokinase 6, beta-D-glucopyranose | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of OsHXK6-Glc complex
To Be Published
|
|
6JJ4
 
 | | Crystal structure of OsHXK6-apo form | | Descriptor: | Hexokinase-6 | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of OsHXK6-apo
To Be Published
|
|
6JJ9
 
 | | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hexokinase-6, MAGNESIUM ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex
To Be Published
|
|
6JJ8
 
 | | Crystal structure of OsHXK6-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of OsHXK6-ATP-Mg2+ complex
To Be Published
|
|
6L6W
 
 | | The structure of ScoE with intermediate | | Descriptor: | (3R)-3-[[(1R)-1,2-bis(oxidanyl)-2-oxidanylidene-ethyl]amino]butanoic acid, FE (II) ION, FORMIC ACID, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8JNX
 
 | |
8JN0
 
 | | N/F domain of alkaline amylase Amy703 | | Descriptor: | Alpha-amylase | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.22826374 Å) | | Cite: | Truncation of N-terminus domain of alkaline a-amylase to form a unique dimer leads to improved activity and stability and decreased calcium ion dependence
To Be Published
|
|
8JLV
 
 | |
7WOI
 
 | | Structure of the shaft pilin Spa2 from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spa2 | | Authors: | Wu, Y.F, Wang, L.T, Huang, Y.Y, Zhong, C, Zhou, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Accelerating the design of pili-enabled living materials using an integrative technological workflow.
Nat.Chem.Biol., 2023
|
|
3TB3
 
 | | Crystal structure of the UCH domain of UCH-L5 with 6 residues deleted | | Descriptor: | CALCIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Zhou, Z.R, Zha, M, Zhou, J, Hu, H.Y. | | Deposit date: | 2011-08-05 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains.
Biochem.J., 441, 2012
|
|
