4E2I
 
 | | The Complex Structure of the SV40 Helicase Large T Antigen and p68 Subunit of DNA Polymerase Alpha-Primase | | Descriptor: | DNA polymerase alpha subunit B, Large T antigen, ZINC ION | | Authors: | Zhou, B, Arnett, D.R, Yu, X, Brewster, A, Sowd, G.A, Xie, C.L, Vila, S, Gai, D, Fanning, E, Chen, X.S. | | Deposit date: | 2012-03-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase.
J.Biol.Chem., 287, 2012
|
|
6KEE
 
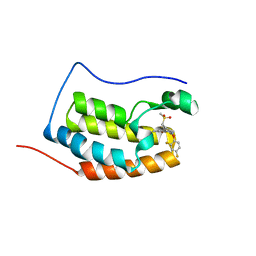 | | Crystal structure of BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12154651 Å) | | Cite: | Crystal structure of BRD4 Bromodomain1 with an inhibitor
To be published
|
|
6KEG
 
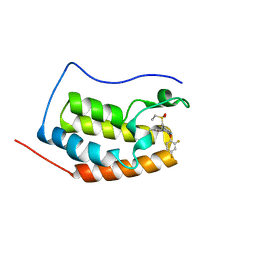 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(ethylsulfonylmethyl)pyridin-3-yl]-8-methyl-4H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KEF
 
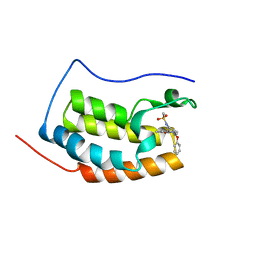 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(8-methyl-1-oxidanylidene-2H-pyrrolo[1,2-a]pyrazin-6-yl)-4-phenoxy-phenyl]methanesulfonamide | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44467545 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KED
 
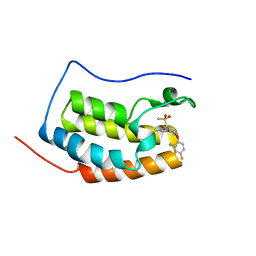 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-d][1,2,4]triazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.548447 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
8T21
 
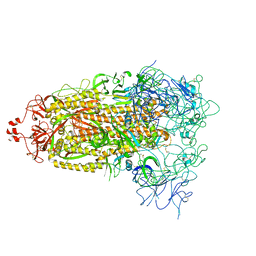 | | Cryo-EM structure of mink variant Y453F trimeric spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T22
 
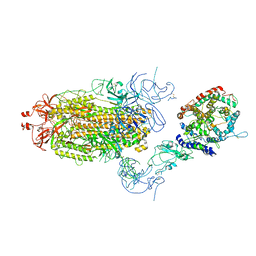 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
3D9C
 
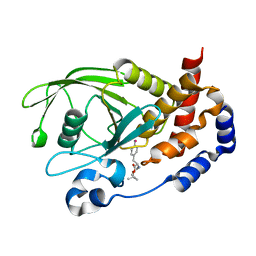 | | Crystal Structure PTP1B complex with aryl Seleninic acid | | Descriptor: | (4-{(2S)-2-[(tert-butoxycarbonyl)amino]-3-methoxy-3-oxopropyl}phenyl)methaneseleninic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Abdo, M, Liu, S, Zhou, B, Walls, C.D, Knapp, S, Zhang, Z.-Y. | | Deposit date: | 2008-05-27 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Seleninate in place of phosphate: irreversible inhibition of protein tyrosine phosphatases.
J.Am.Chem.Soc., 130, 2008
|
|
3HF1
 
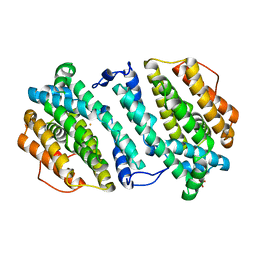 | | Crystal structure of human p53R2 | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase subunit M2 B, SULFATE ION | | Authors: | Smith, P, Zhou, B, Yuan, Y.-C, Su, L, Tsai, S.-C, Yen, Y. | | Deposit date: | 2009-05-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A X-ray crystal structure of human p53R2, a p53-inducible ribonucleotide reductase .
Biochemistry, 48, 2009
|
|
4HQ1
 
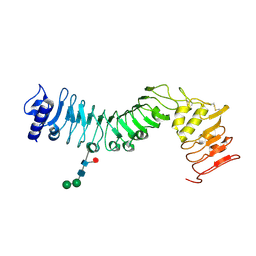 | | Crystal structure of an LRR protein with two solenoids | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Probable receptor protein kinase TMK1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chai, J, Liu, P, Hu, Z, Zhou, B, Liu, S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of an LRR protein with two solenoids
Cell Res., 23, 2013
|
|
4J0M
 
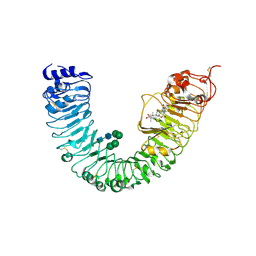 | | Crystal structure of BRL1 (LRR) in complex with brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, ... | | Authors: | Chai, J, She, J, Han, Z, Zhou, B. | | Deposit date: | 2013-01-31 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for differential recognition of brassinolide by its receptors
Protein Cell, 4, 2013
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
2FYS
 
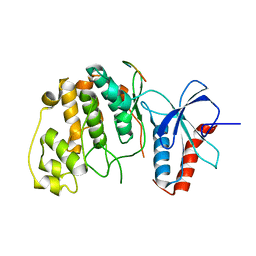 | | Crystal structure of Erk2 complex with KIM peptide derived from MKP3 | | Descriptor: | Dual specificity protein phosphatase 6, Mitogen-activated protein kinase 1 | | Authors: | Liu, S, Sun, J.P, Zhou, B, Zhang, Z.Y. | | Deposit date: | 2006-02-08 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
2L5A
 
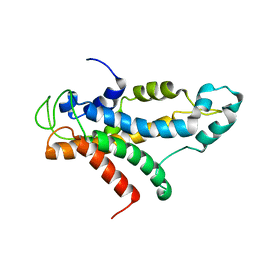 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
8GJM
 
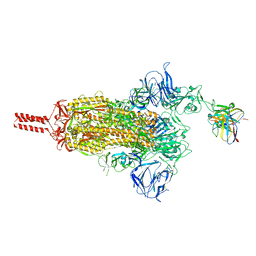 | | 17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8GJN
 
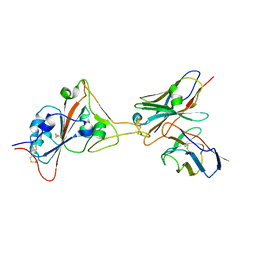 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
5DAB
 
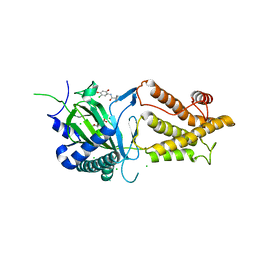 | | Crystal structure of FTO-IN115 | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, CHLORIDE ION, ... | | Authors: | Chai, J, Zhou, B, Liu, W, Han, Z, Niu, T. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of FTO-IN115
To Be Published
|
|
5F8P
 
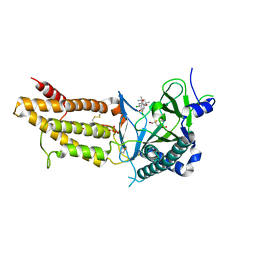 | | A Novel Inhibitor of the Obesity-Related Protein FTO | | Descriptor: | 2-OXOGLUTARIC ACID, 4-chloranyl-6-[(2~{S})-6-chloranyl-2,4,4-trimethyl-7-oxidanyl-3~{H}-chromen-2-yl]benzene-1,3-diol, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Chai, J, Zhou, B, Liu, W, Han, Z. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FTO-CHTB
To Be Published
|
|
2LP0
 
 | |
5YEY
 
 | |
8YHX
 
 | | Cryo-EM structure of the trimeric HerA | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, DUF87 domain-containing protein, SIR2 family protein | | Authors: | Zhen, X, Zhou, B, Xiong, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Mechanistic basis for the allosteric activation of NADase activity in the Sir2-HerA antiphage defense system.
Nat Commun, 15, 2024
|
|
8T25
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation. | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T23
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
