5UIW
 
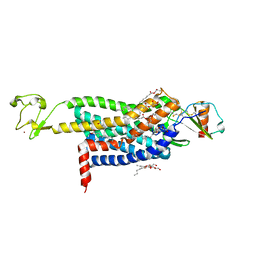 | | Crystal Structure of CC Chemokine Receptor 5 (CCR5) in complex with high potency HIV entry inhibitor 5P7-CCL5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, C-C chemokine receptor type 5,Rubredoxin chimera, C-C motif chemokine 5, ... | | Authors: | Zheng, Y, Qin, L, Han, G.W, Gustavsson, M, Kawamura, T, Stevens, R.C, Cherezov, V, Kufareva, I, Handel, T.M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structure of CC Chemokine Receptor 5 with a Potent Chemokine Antagonist Reveals Mechanisms of Chemokine Recognition and Molecular Mimicry by HIV.
Immunity, 46, 2017
|
|
5XWZ
 
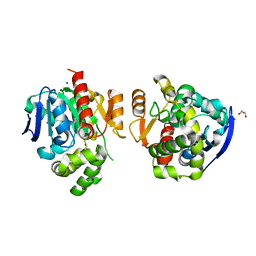 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3WGG
 
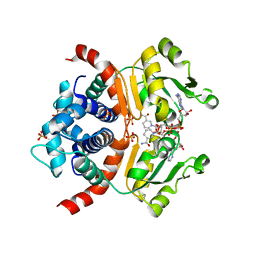 | | Crystal structure of RSP in complex with alpha-NAD+ | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION, alpha-Diphosphopyridine nucleotide | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding mode of the oxidized alpha-anomer of NAD(+) to RSP, a Rex-family repressor
Biochem.Biophys.Res.Commun., 456, 2015
|
|
3X2E
 
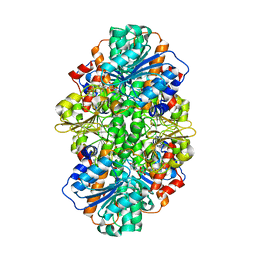 | | A thermophilic hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
6URQ
 
 | | Complex structure of human poly-ADP-ribosyltransferase TNKS1 ARC2-ARC3 and P antigen family member 4 (PAGE4) | | Descriptor: | GLYCEROL, P antigen family member 4, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Zheng, Y, Koirala, S, Miller, D, Potts, P.R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tissue-Specific Regulation of the Wnt/ beta-Catenin Pathway by PAGE4 Inhibition of Tankyrase.
Cell Rep, 32, 2020
|
|
6OJJ
 
 | |
3X2F
 
 | | A Thermophilic S-Adenosylhomocysteine Hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase, NITRATE ION, ... | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
7JH4
 
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
3VMV
 
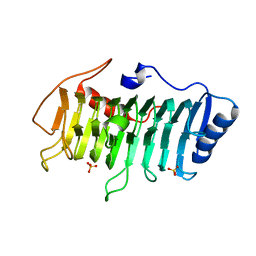 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
3VMW
 
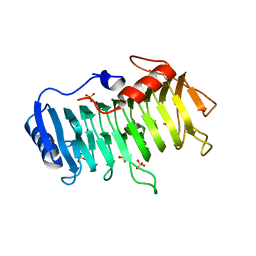 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate | | Descriptor: | Pectate lyase, SULFATE ION, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
4YIA
 
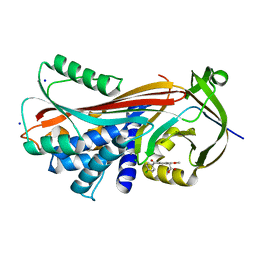 | |
6V99
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase- S72D in the presence of sulfate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V96
 
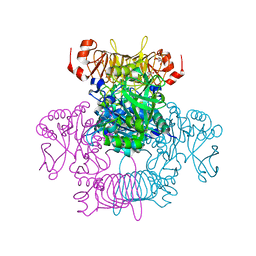 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72E | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Hussien, R, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V9A
 
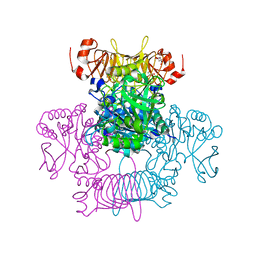 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72D | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6JBY
 
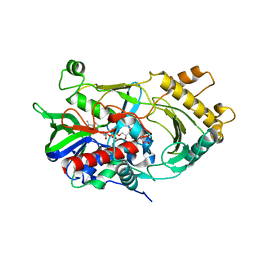 | | Crystal structure of endo-deglycosylated hydroxynitrile lyase isozyme 5 of Prunus communis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2019-01-27 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
5Z97
 
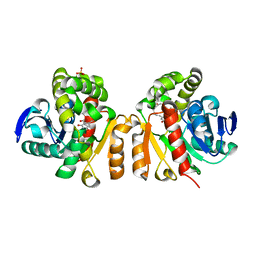 | | Crystal structure of a lactonase double mutant in complex with ligand N | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein, SULFATE ION | | Authors: | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-02-02 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5XO6
 
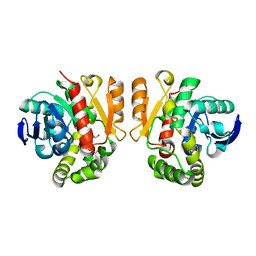 | | Crystal structure of a novel ZEN lactonase mutant | | Descriptor: | PENTAETHYLENE GLYCOL, lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5XO7
 
 | | Crystal structure of a novel ZEN lactonase mutant with ligand a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5Z5J
 
 | | Crystal structure of a lactonase double mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5XO8
 
 | | Crystal structure of a novel ZEN lactonase mutant with ligand Z | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5Z7J
 
 | | Crystal structure of a lactonase double mutant in complex with ligand l | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, DI(HYDROXYETHYL)ETHER, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
7U58
 
 | |
7VV9
 
 | |
7VVG
 
 | |
7VVB
 
 | |
