4Q4F
 
 | | Crystal structure of LIMP-2 (space group C2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
8JCO
 
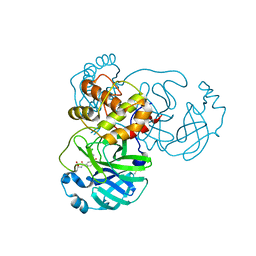 | |
8JCL
 
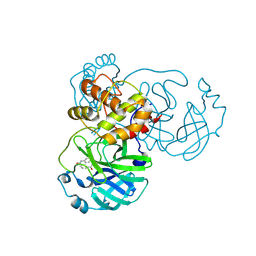 | |
8JCK
 
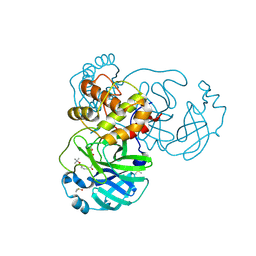 | |
8JCM
 
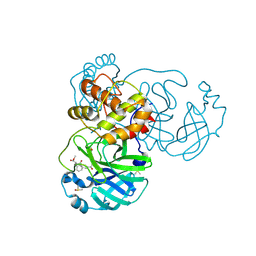 | |
8JCJ
 
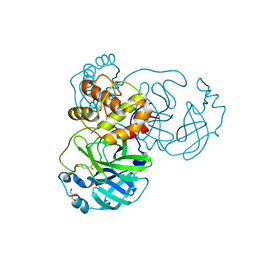 | |
8JCN
 
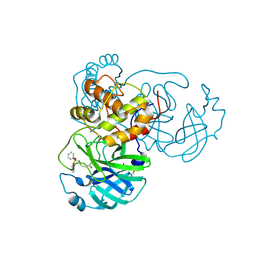 | |
4Q4B
 
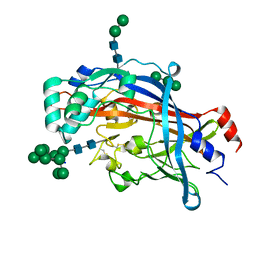 | | Crystal structure of LIMP-2 (space group C2221) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
8EVT
 
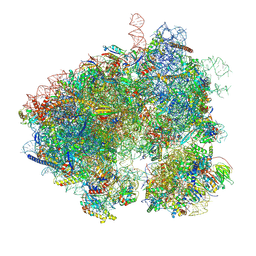 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES) refined against a composite map | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVR
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure II | | Descriptor: | (1S,3S,3aR,4S,4aR,7R,7aR,8aS)-8a-{[(6-deoxy-4-O-methyl-alpha-D-altropyranosyl)oxy]methyl}-4-formyl-7-methyl-3-(propan-2-yl)decahydro-1,4-methano-s-indacene-3a(1H)-carboxylate, 18S rRNA, 25S rRNA, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVS
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2 and GDP, Structure II | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVQ
 
 | | Hypopseudouridylated Ribosome bound with TSV IRES, eEF2, GDP, and sordarin, Structure I | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EWC
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), Structure II | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EWB
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2 and GDP, Structure III | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
1VBS
 
 | | STRUCTURE OF CYCLOPHILIN COMPLEXED WITH (D)ALA CONTAINING TETRAPEPTIDE | | Descriptor: | CYCLOPHILIN A, TETRAPEPTIDE | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the stereospecificity of peptidyl prolyl cis/trans isomerases.
FEBS Lett., 432, 1998
|
|
6JP8
 
 | | Rabbit Cav1.1-Bay K8644 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6JPB
 
 | | Rabbit Cav1.1-Diltiazem Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6JPA
 
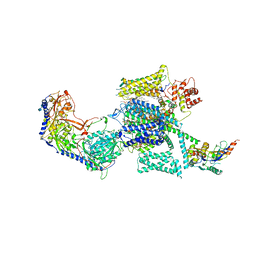 | | Rabbit Cav1.1-Verapamil Complex | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6JP5
 
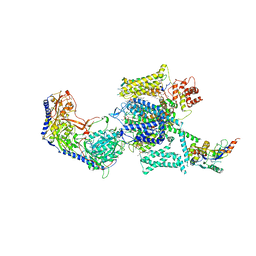 | | Rabbit Cav1.1-Nifedipine Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7X31
 
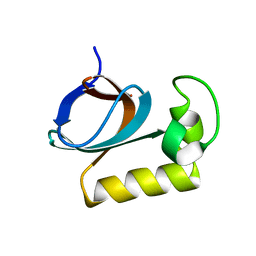 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7W33
 
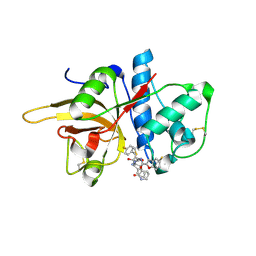 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
7W7O
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14a | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2021-12-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14a
To Be Published
|
|
