8JGP
 
 | |
8JGO
 
 | |
8Y2C
 
 | | Cryo-EM structure of human dopamine transporter in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium-dependent dopamine transporter | | Authors: | Zhao, Y, Li, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Dopamine reuptake and inhibitory mechanisms in human dopamine transporter.
Nature, 632, 2024
|
|
8Y2E
 
 | |
8Y2G
 
 | |
8Y2F
 
 | | Cryo-EM structure of human dopamine transporter in complex with GBR12909 | | Descriptor: | 1-[2-[bis(4-fluorophenyl)methoxy]ethyl]-4-(3-phenylpropyl)piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium-dependent dopamine transporter | | Authors: | Zhao, Y, Li, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Dopamine reuptake and inhibitory mechanisms in human dopamine transporter.
Nature, 632, 2024
|
|
8Y2D
 
 | |
5WSZ
 
 | | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis | | Descriptor: | COPPER (II) ION, LpmO10A | | Authors: | Zhao, Y, Zhang, H, Yin, H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis
To Be Published
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
3CYH
 
 | | CYCLOPHILIN A COMPLEXED WITH DIPEPTIDE SER-PRO | | Descriptor: | CYCLOPHILIN A, PROLINE, SERINE | | Authors: | Zhao, Y, Ke, H. | | Deposit date: | 1996-02-27 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic implication of crystal structures of the cyclophilin-dipeptide complexes.
Biochemistry, 35, 1996
|
|
6TR6
 
 | | N-acetylserotonin-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
6TV4
 
 | | CFF-Notum complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
6TUZ
 
 | | Theophylline-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
8YZ7
 
 | |
4YVP
 
 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVV
 
 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
5VOV
 
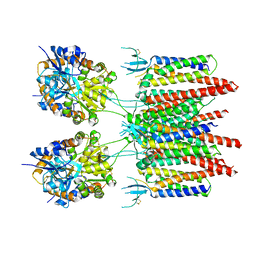 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
4ZFC
 
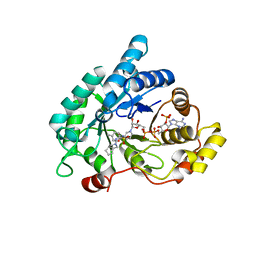 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
7S1Y
 
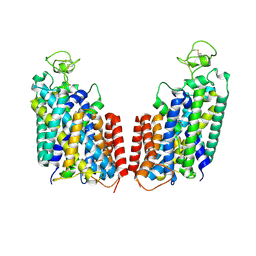 | | Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanide | | Descriptor: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Zhao, Y.X, Cao, E.h. | | Deposit date: | 2021-09-02 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for inhibition of the Cation-chloride cotransporter NKCC1 by the diuretic drug bumetanide
Nat Commun, 13, 2022
|
|
7S1X
 
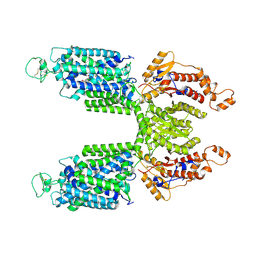 | |
7S1Z
 
 | | Cryo-EM structure of Human NKCC1 K289NA492EL671C | | Descriptor: | Solute carrier family 12 member 2 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2021-09-02 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for inhibition of the Cation-chloride cotransporter NKCC1 by the diuretic drug bumetanide
Nat Commun, 13, 2022
|
|
7QVZ
 
 | | ARUK3001043_Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Hillier, J, Willis, N.J, Mahy, W, Fish, P, Jones, E.Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure Activity Analysis of Notum Fragment Screen Hits with Development
To Be Published
|
|
7B8U
 
 | | Notum-Fragment 201 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(piperidin-1-yl)-1,2,5-oxadiazol-3-amine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8N
 
 | | Notum-Fragment 197 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-azanyl-2-(pyridin-3-ylmethylamino)benzoic acid, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
