1I7U
 
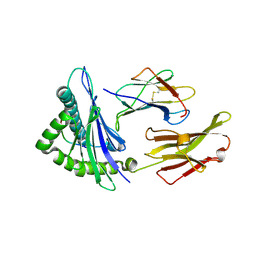 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1049-6V | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | T cell activity correlates with oligomeric peptide-major histocompatibility complex binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
3N3Z
 
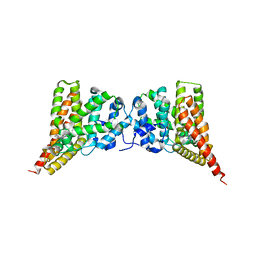 | | Crystal structure of PDE9A (E406A) mutant in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CHLORIDE ION, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Hou, J, Luo, H.-B, Chen, Y, Xu, J, Zhao, R, Zou, L. | | Deposit date: | 2010-05-21 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PDE9A (E406A) mutation in complex with IBMX
To be Published
|
|
1OOI
 
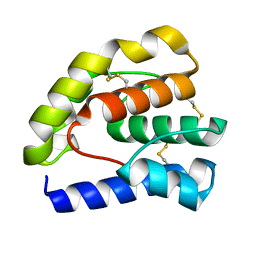 | |
1OOH
 
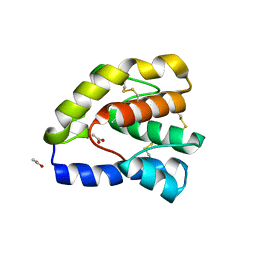 | | Complex of Drosophila odorant binding protein LUSH with butanol | | Descriptor: | 1-BUTANOL, ACETATE ION, odorant binding protein LUSH | | Authors: | Kruse, S.W, Zhao, R, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2003-03-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster
Nat.Struct.Biol., 10, 2003
|
|
1OOF
 
 | | Complex of Drosophila odorant binding protein LUSH with ethanol | | Descriptor: | ACETATE ION, ETHANOL, odorant binding protein LUSH | | Authors: | Kruse, S.W, Zhao, R, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2003-03-03 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster
Nat.Struct.Biol., 10, 2003
|
|
1OOG
 
 | | Complex of Drosophila odorant binding protein LUSH with propanol | | Descriptor: | ACETATE ION, N-PROPANOL, odorant binding protein LUSH | | Authors: | Kruse, S.W, Zhao, R, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2003-03-03 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster
Nat.Struct.Biol., 10, 2003
|
|
1MIH
 
 | | A ROLE FOR CHEY GLU 89 IN CHEZ-MEDIATED DEPHOSPHORYLATION OF THE E. COLI CHEMOTAXIS RESPONSE REGULATOR CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, MANGANESE (II) ION, ... | | Authors: | Silversmith, R.E, Guanga, G.P, Betts, L, Chu, C, Zhao, R, Bourret, R.B. | | Deposit date: | 2002-08-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89.
J.Bacteriol., 185, 2003
|
|
1D4Z
 
 | | CRYSTAL STRUCTURE OF CHEY-95IV, A HYPERACTIVE CHEY MUTANT | | Descriptor: | CHEMOTAXIS PROTEIN CHEY, SULFATE ION | | Authors: | Schuster, M, Zhao, R, Bourret, R.B, Collins, E.J. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlated switch binding and signaling in bacterial chemotaxis.
J.Biol.Chem., 275, 2000
|
|
8JED
 
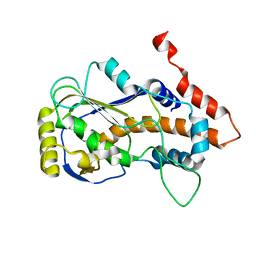 | | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors | | Descriptor: | mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Wang, D, Zhao, R, Shu, W, Hu, W, Wang, M, Cao, J, Zhou, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors.
Int.J.Biol.Macromol., 253, 2023
|
|
1N25
 
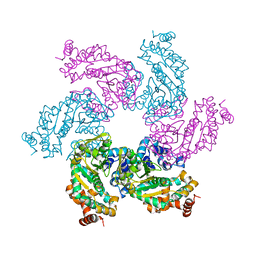 | | Crystal structure of the SV40 Large T antigen helicase domain | | Descriptor: | Large T Antigen, ZINC ION | | Authors: | Li, D, Zhao, R, Lilyestrom, W, Gai, D, Zhang, R, DeCaprio, J.A, Fanning, E, Jochimiak, A, Szakonyi, G, Chen, X.S. | | Deposit date: | 2002-10-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature, 423, 2003
|
|
6N7P
 
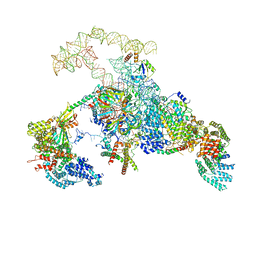 | | S. cerevisiae spliceosomal E complex (UBC4) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Nuclear cap-binding protein complex subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
6N7X
 
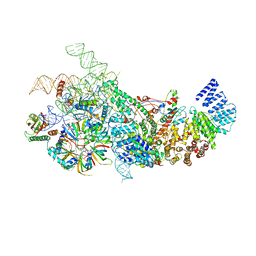 | | S. cerevisiae U1 snRNP | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | Authors: | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
1SVM
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
6N7R
 
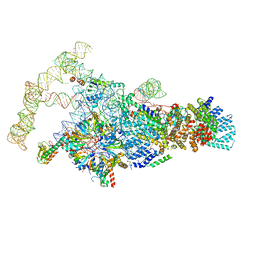 | | Saccharomyces cerevisiae spliceosomal E complex (ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
1SVL
 
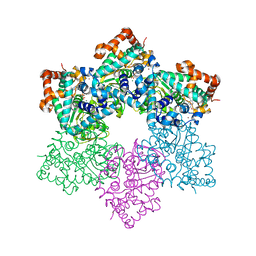 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVO
 
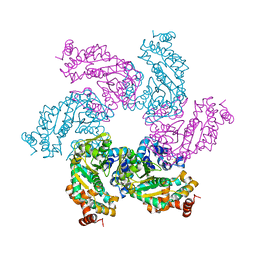 | | Structure of SV40 large T antigen helicase domain | | Descriptor: | ZINC ION, large T antigen | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
8W2O
 
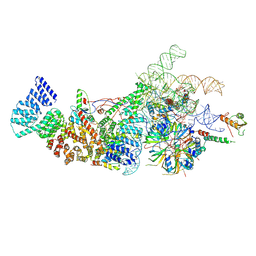 | | Yeast U1 snRNP with humanized U1C Zinc-Finger domain | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | Authors: | Shi, S.S, Kuang, Z.L, Zhao, R. | | Deposit date: | 2024-02-20 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Selected humanization of yeast U1 snRNP leads to global suppression of pre-mRNA splicing and mitochondrial dysfunction in the budding yeast.
Rna, 30, 2024
|
|
6BK8
 
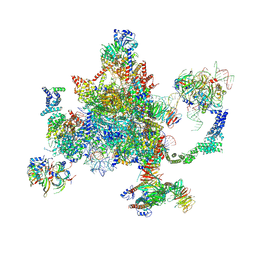 | | S. cerevisiae spliceosomal post-catalytic P complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
2G6Q
 
 | |
5GVW
 
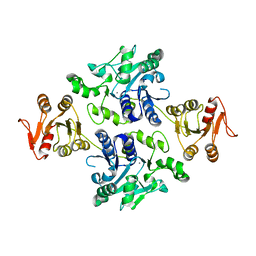 | | Crystal structure of the apo-form glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
5GVV
 
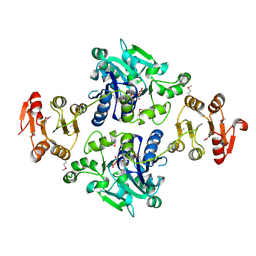 | | Crystal structure of the glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
7EPT
 
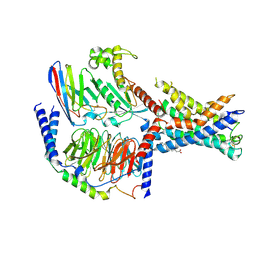 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
2L6J
 
 | | Tah1 complexed by MEEVD | | Descriptor: | C-terminus Hsp90 chaperone peptide MEEVD, TPR repeat-containing protein associated with Hsp90 | | Authors: | Jimenez, B, Ugwu, F, Zhao, R, Orti, L, Houry, W.A, Pineda-Lucena, A. | | Deposit date: | 2010-11-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90.
J.Biol.Chem., 287, 2012
|
|
2N6B
 
 | |
2P8R
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 carrying R2303K mutant | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
