3U3T
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3V
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8YW5
 
 | | Cryo-EM structure of the retatrutide-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
2RHK
 
 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | Authors: | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BHP
 
 | | Crystal structure of UPF0291 protein ynzC from Bacillus subtilis at resolution 2.0 A. Northeast Structural Genomics Consortium target SR384 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Janjua, H, Cunningham, K, Maglaqui, M, Owens, L.A, Zhao, L, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the UPF0291 protein ynzC from Bacillus subtilis at the resolution 2.0 A.
To be Published
|
|
4FIB
 
 | | Crystal structure of the ydhK protein from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR518A. | | Descriptor: | Uncharacterized protein ydhK | | Authors: | Vorobiev, S, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Maglaqui, M, Owens, L.A, Cunningham, K, Zhao, L, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of the ydhK protein from Bacillus subtilis.
To be Published
|
|
6PB0
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6EWS
 
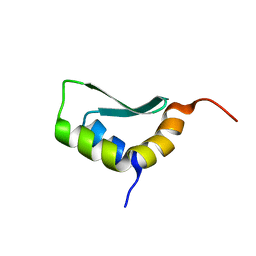 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWV
 
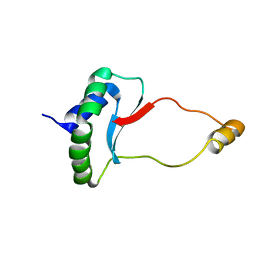 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | Descriptor: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6PB1
 
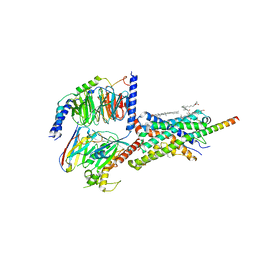 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
4DLH
 
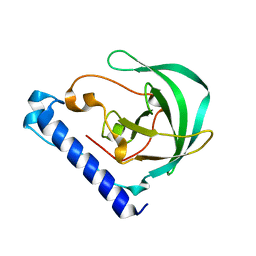 | | Crystal Structure of the protein Q9HRE7 from Halobacterium salinarium at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HsR50 | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
7BRE
 
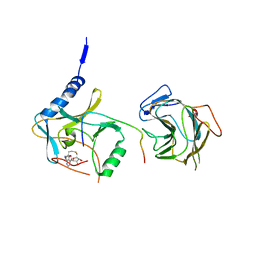 | | The crystal structure of MLL2 in complex with ASH2L and RBBP5 | | Descriptor: | Histone-lysine N-methyltransferase 2B, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Zhao, L, Chen, Y. | | Deposit date: | 2020-03-28 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal Structure of MLL2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by KMT2 Family Methyltransferases.
Structure, 28, 2020
|
|
6EWT
 
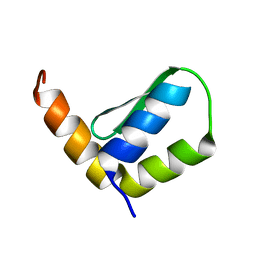 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWU
 
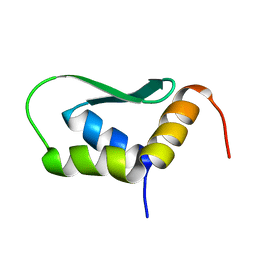 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
5L03
 
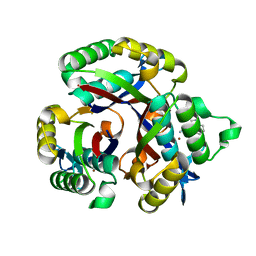 | | Crystal structure of 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE Synthase from BURKHOLDERIA PSEUDOMALLEI bound to L-tryptophan hydroxamate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, N-hydroxy-L-tryptophanamide, ZINC ION | | Authors: | Blain, J.M, Ghose, D, Gorman, J.L, Goshu, G.M, Ranieri, G, Zhao, L, Bode, B, Meganathan, R, Walter, R.L, Hagen, T.J, Horn, J.R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Synthesis and Characterization of the Burkholderia pseudomallei IspF Inhibitor L-tryptophan hydroxamate
To Be Published
|
|
6NHW
 
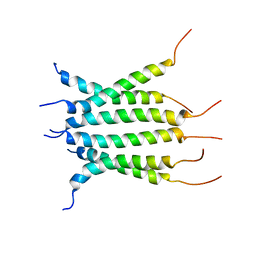 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
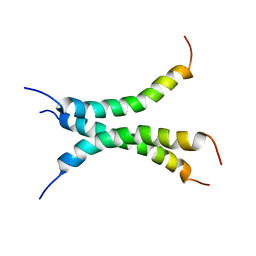 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
5DJ5
 
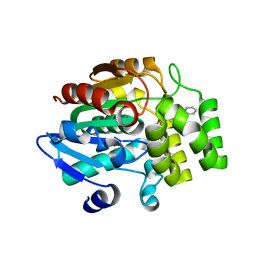 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
3MEL
 
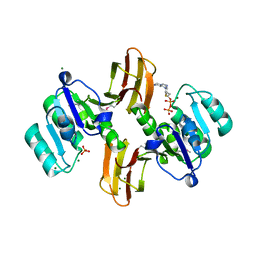 | | Crystal Structure of Thiamin pyrophosphokinase family protein from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR150 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Kuzin, A, Abasidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Northeast Structural Genomics Consortium Target EfR150
To be Published
|
|
6K40
 
 | | Crystal structure of alkyl hydroperoxide reductase from D. radiodurans R1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase AhpD, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, M.-K, Zhang, J, Zhao, L. | | Deposit date: | 2019-05-22 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the AhpD-like protein DR1765 from Deinococcus radiodurans R1.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
3MGD
 
 | | Crystal Structure of predicted acetyltransferase with acetyl-CoA from Clostridium acetobutylicum at the resolution 1.9A, Northeast Structural Genomics Consortium Target CaR165 | | Descriptor: | ACETYL COENZYME *A, Predicted acetyltransferase | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-28 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target CaR165
To be published
|
|
6IZ2
 
 | |
4QGU
 
 | | protein domain complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | Authors: | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | Deposit date: | 2014-05-25 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
4NFZ
 
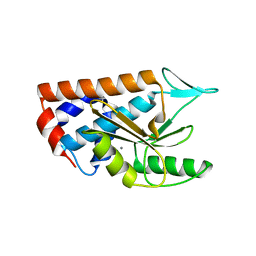 | | Crystal structure of polymerase subunit PA N-terminal endonuclease domain from bat-derived influenza virus H17N10 | | Descriptor: | MANGANESE (II) ION, Polymerase PA | | Authors: | Tefsen, B, Lu, G, Zhu, Y, Haywood, J, Zhao, L, Deng, T, Qi, J, Gao, G.F. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N-Terminal Domain of PA from Bat-Derived Influenza-Like Virus H17N10 Has Endonuclease Activity
J.Virol., 88, 2014
|
|
3MQP
 
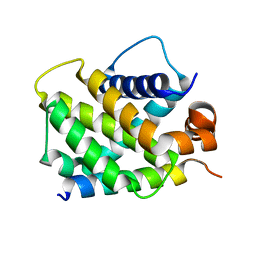 | | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide, Northeast Structural Genomics Consortium Target HR2930 | | Descriptor: | Bcl-2-related protein A1, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Guan, R, Xiao, R, Zhao, L, Acton, T.B, Gelinas, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide
To be Published
|
|
