7KPT
 
 | | Crystal structure of CtdE in complex with FAD and substrate 4 | | Descriptor: | (6aR,7aS,11S,13aS)-6,6,11-trimethyl-4-(3-methylbut-2-en-1-yl)-6,6a,7,8,9,10,11,14-octahydro-5H,13H-13a,7a-(epiminomethano)quinolizino[2,3-b]carbazol-16-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7KPQ
 
 | | Crystal structure of CtdE in complex with FAD | | Descriptor: | FAD-dependent monooxygenase CtdE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
1NVQ
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVR
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVS
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/SB218078 | | Descriptor: | Peptide ASVSA, REL-(9R,12S)-9,10,11,12-TETRAHYDRO-9,12-EPOXY-1H-DIINDOLO[1,2,3-FG:3',2',1'-KL]PYRROLO[3,4-I][1,6]BENZODIAZOCINE-1,3(2H)-DIONE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
3HA6
 
 | | Crystal structure of aurora A in complex with TPX2 and compound 10 | | Descriptor: | N~2~-(3,4-dimethoxyphenyl)-N~4~-[2-(2-fluorophenyl)ethyl]-N~6~-quinolin-6-yl-1,3,5-triazine-2,4,6-triamine, Serine/threonine-protein kinase 6, Targeting protein for Xklp2 | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
6V7O
 
 | |
6UOZ
 
 | |
5UJB
 
 | | Structure of a Mcl-1 Inhibitor Binding to Site 3 of Human Serum Albumin | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, PHOSPHATE ION, Serum albumin | | Authors: | Zhao, B. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Myeloid cell leukemia-1 (Mcl-1) inhibitor bound to drug site 3 of Human Serum Albumin.
Bioorg. Med. Chem., 25, 2017
|
|
3PDF
 
 | | Discovery of Novel Cyanamide-Based Inhibitors of Cathepsin C | | Descriptor: | 2,5-dibromo-N-{(3R,5S)-1-[(Z)-iminomethyl]-5-methylpyrrolidin-3-yl}benzenesulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, B, Laine, D. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of novel cyanamide-based inhibitors of cathepsin C.
Acs Med.Chem.Lett., 2, 2011
|
|
5IF4
 
 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
5IEZ
 
 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 3-({6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-1H-indole-2-carbonyl}amino)benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
5JEK
 
 | | Phosphorylated MAVS in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, MAVS peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEM
 
 | | Complex of IRF-3 with CBP | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JER
 
 | | Structure of Rotavirus NSP1 bound to IRF-3 | | Descriptor: | Interferon regulatory factor 3, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEL
 
 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5U0B
 
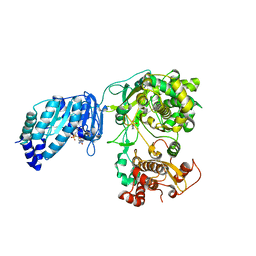 | | Structure of full-length Zika virus NS5 | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhao, B, Du, F. | | Deposit date: | 2016-11-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the Zika virus full-length NS5 protein.
Nat Commun, 8, 2017
|
|
5U0C
 
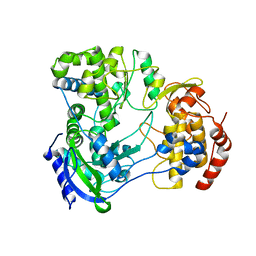 | |
7JJ1
 
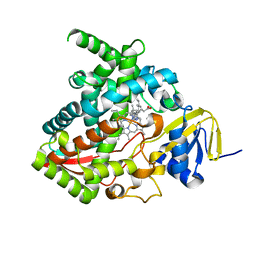 | |
4ZBI
 
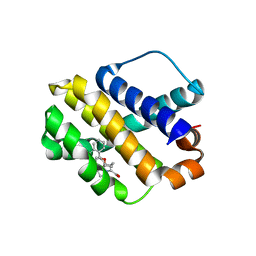 | | Mcl-1 complexed with small molecules | | Descriptor: | 1-[3-(naphthalen-1-yloxy)propyl]-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
4ZBF
 
 | | Mcl-1 complexed with small molecules | | Descriptor: | (1R)-7-[3-(naphthalen-1-yloxy)propyl]-3,4-dihydro-2H-[1,4]thiazepino[2,3,4-hi]indole-6-carboxylic acid 1-oxide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
3DBG
 
 | |
3EL3
 
 | | Distinct Monooxygenase and Farnesene Synthase Active Sites in Cytochrome P450 170A1 | | Descriptor: | (3S,3aR,6S)-3,7,7,8-tetramethyl-2,3,4,5,6,7-hexahydro-1H-3a,6-methanoazulene, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site
J.Biol.Chem., 284, 2009
|
|
3E5A
 
 | | Crystal structure of Aurora A in complex with VX-680 and TPX2 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, SULFATE ION, Serine/threonine-protein kinase 6, ... | | Authors: | Zhao, B, Smallwood, A, Lai, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2.
Protein Sci., 17, 2008
|
|
