2OPW
 
 | | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo) | | Descriptor: | PHYHD1 protein | | Authors: | Zhang, Z, Butler, D, McDonough, M.A, Kavanagh, K.L, Bray, J.E, Ng, S.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo)
To be Published
|
|
5UAR
 
 | |
5W81
 
 | | Phosphorylated, ATP-bound structure of zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2017-06-21 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational Changes of CFTR upon Phosphorylation and ATP Binding.
Cell, 170, 2017
|
|
5GMG
 
 | | Crystal structure of monkey TLR7 in complex with loxoribine and polyU | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-7-prop-2-enyl-1~{H}-purine-6,8-dione, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA
Immunity, 45, 2016
|
|
5GMH
 
 | | Crystal structure of monkey TLR7 in complex with R848 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA
Immunity, 45, 2016
|
|
5H6V
 
 | |
5GMF
 
 | | Crystal structure of monkey TLR7 in complex with guanosine and polyU | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA
Immunity, 45, 2016
|
|
8GJK
 
 | | Multi-drug efflux pump RE-CmeB bound with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJL
 
 | | multi-drug efflux pump RE-CmeB bound with Ciprofloxacin | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJJ
 
 | | Multi-drug efflux pump RE-CmeB Apo form | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GK4
 
 | | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GK0
 
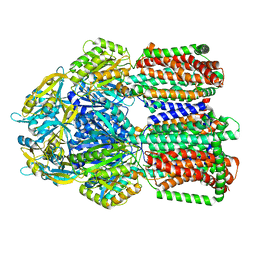 | | Multi-drug efflux pump RE-CmeB bound with Erythromycin | | Descriptor: | ERYTHROMYCIN A, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8UVK
 
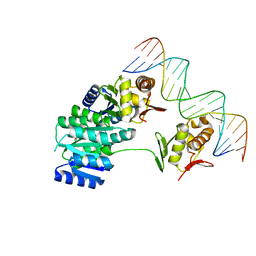 | | CosR DNA bound form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
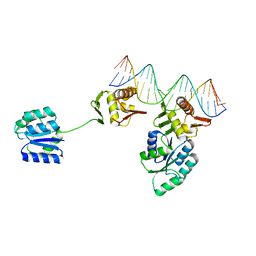 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
2JX0
 
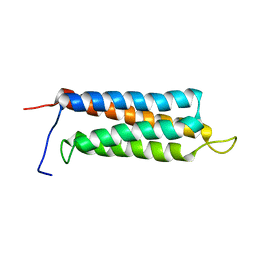 | | The paxillin-binding domain (PBD) of G Protein Coupled Receptor (GPCR)-kinase (GRK) interacting protein 1 (GIT1) | | Descriptor: | ARF GTPase-activating protein GIT1 | | Authors: | Zhang, Z, Guibao, C.D, Simmerman, J.A, Zheng, J. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs.
J.Biol.Chem., 283, 2008
|
|
3U9I
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Roseiflexus sp. | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, C-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-19 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Roseiflexus sp.
TO BE PUBLISHED
|
|
3V5N
 
 | | The crystal structure of oxidoreductase from Sinorhizobium meliloti | | Descriptor: | Oxidoreductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-16 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | The crystal structure of oxidoreductase from Sinorhizobium meliloti
To be Published
|
|
3SSZ
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium
To be Published
|
|
3T4W
 
 | | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp
To be Published
|
|
3T7U
 
 | | A NeW Crystal structure of APC-ARM | | Descriptor: | Adenomatous polyposis coli protein, PHOSPHATE ION | | Authors: | Zhang, Z, Wu, G. | | Deposit date: | 2011-07-31 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the armadillo repeat domain of adenomatous polyposis coli which reveals its inherent flexibility
Biochem.Biophys.Res.Commun., 412, 2011
|
|
7E3J
 
 | |
3VA9
 
 | |
3TG9
 
 | | The crystal structure of penicillin binding protein from Bacillus halodurans | | Descriptor: | Penicillin-binding protein | | Authors: | Zhang, Z, Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of penicillin binding protein from Bacillus halodurans
To be Published
|
|
3V9F
 
 | |
