3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5CG9
 
 | | NgTET1 in complex with 5mC DNA in space group P3221 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CG8
 
 | | NgTET1 in complex with 5hmC DNA | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5EH2
 
 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
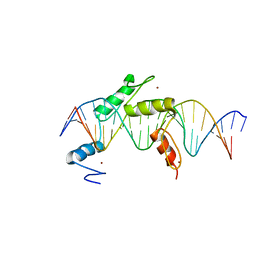 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EGB
 
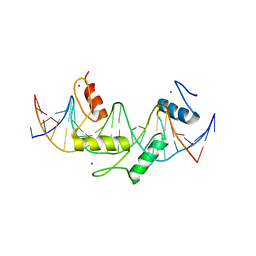 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
3KV4
 
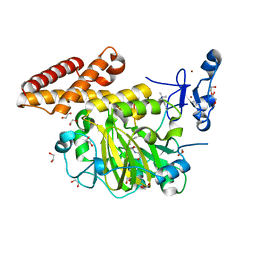 | | Structure of PHF8 in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVB
 
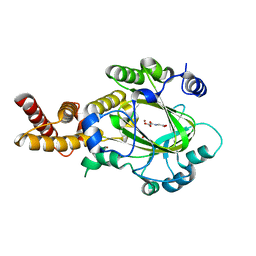 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3J6C
 
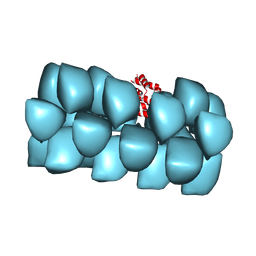 | | Cryo-EM structure of MAVS CARD filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | Deposit date: | 2014-02-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1R7R
 
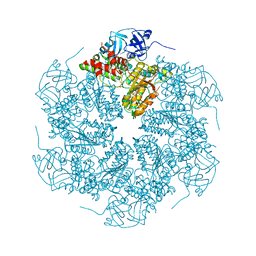 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
1S3S
 
 | | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin containing protein) (VCP) [Contains: Valosin], p47 protein | | Authors: | Dreveny, I, Kondo, H, Uchiyama, K, Shaw, A, Zhang, X, Freemont, P.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47.
Embo J., 23, 2004
|
|
1RHP
 
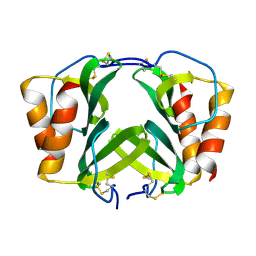 | |
5FB1
 
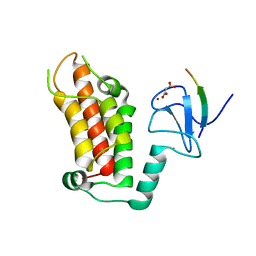 | | Crystal Structure of a PHD finger bound to histone H3 K9me3 peptide | | Descriptor: | MALONATE ION, Nuclear autoantigen Sp-100, Peptide from Histone H3, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
1WOV
 
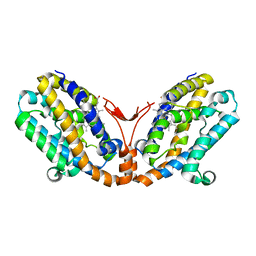 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
1WOW
 
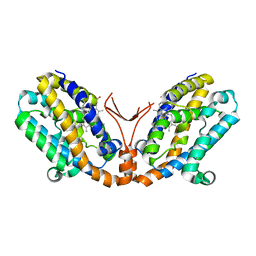 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 complexed with heme in ferrous form | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
8BNZ
 
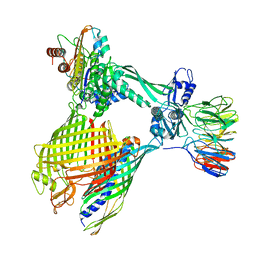 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
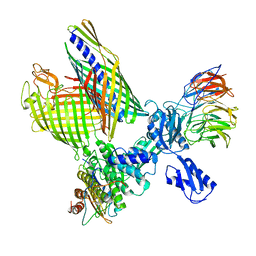 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
1WOX
 
 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme and NO | | Descriptor: | Heme oxygenase 2, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
7ARM
 
 | | LolCDE in complex with lipoprotein and LolA | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARL
 
 | | LolCDE in complex with lipoprotein and ADP | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, ADENOSINE-5'-DIPHOSPHATE, LPP, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARJ
 
 | | LolCDE in complex with lipoprotein and AMPPNP complex undimerized form | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARI
 
 | | LolCDE apo structure | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARK
 
 | | LolCDE in complex with AMP-PNP in the closed NBD state | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
