5CG9
 
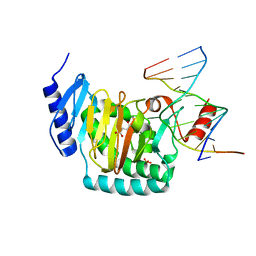 | | NgTET1 in complex with 5mC DNA in space group P3221 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CG8
 
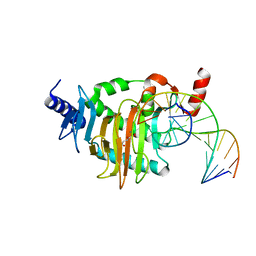 | | NgTET1 in complex with 5hmC DNA | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
2IPA
 
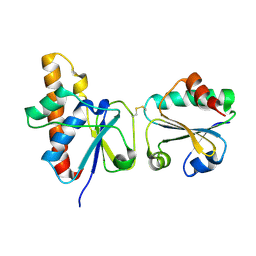 | | solution structure of Trx-ArsC complex | | Descriptor: | Protein arsC, Thioredoxin | | Authors: | Jin, C, Hu, Y, Li, Y, Zhang, X. | | Deposit date: | 2006-10-12 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1U2Z
 
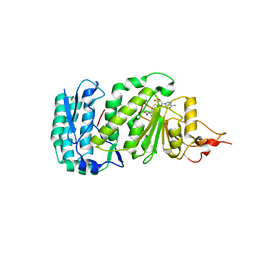 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
5EH2
 
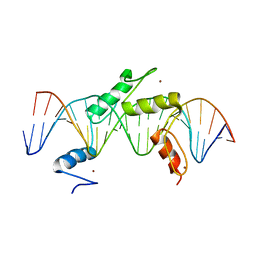 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
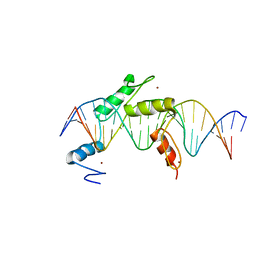 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EGB
 
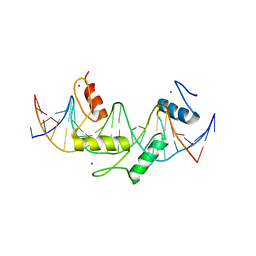 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
3UT7
 
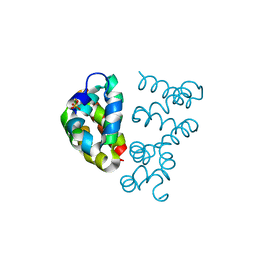 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3KXI
 
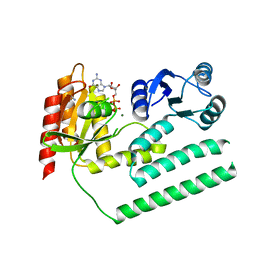 | | crystal structure of SsGBP and GDP complex | | Descriptor: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KV4
 
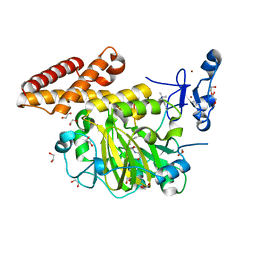 | | Structure of PHF8 in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4ZDS
 
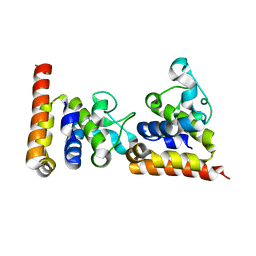 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
3M95
 
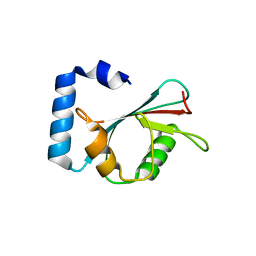 | | Crystal structure of autophagy-related protein Atg8 from the silkworm Bombyx mori | | Descriptor: | Autophagy related protein Atg8 | | Authors: | Teng, Y.-B, Hu, C, Zhang, X, Jiang, Y.L, Hu, H.-X, Zhou, C.Z. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of autophagy-related protein Atg8 from the silkworm Bombyx mori
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2B37
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-octyl-2-phenoxyphenol | | Descriptor: | 5-OCTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3S8M
 
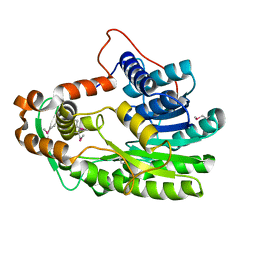 | | The Crystal Structure of FabV | | Descriptor: | Enoyl-ACP Reductase | | Authors: | Li, H, Zhang, X.L, Bi, L.J, He, J, Jiang, T. | | Deposit date: | 2011-05-29 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determination of the Crystal Structure and Active Residues of FabV, the Enoyl-ACP Reductase from Xanthomonas oryzae.
Plos One, 6, 2011
|
|
3KV5
 
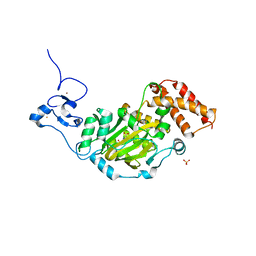 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV9
 
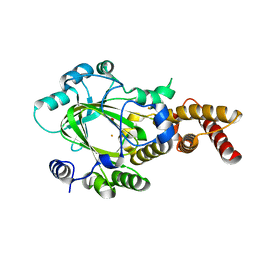 | | Structure of KIAA1718 Jumonji domain | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8KI4
 
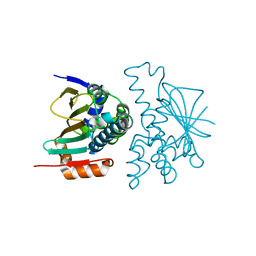 | | Crystal structure of human HSP90 in intermediate state | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein HSP 90-alpha | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-08-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
7XQP
 
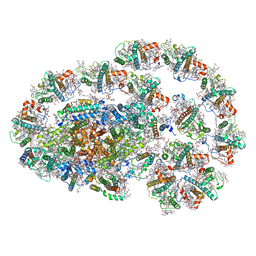 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
3KVA
 
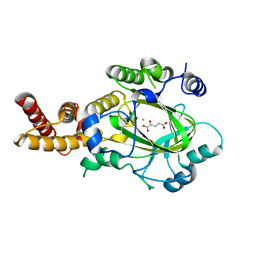 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV6
 
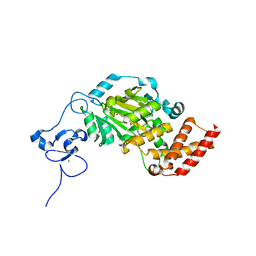 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4IP3
 
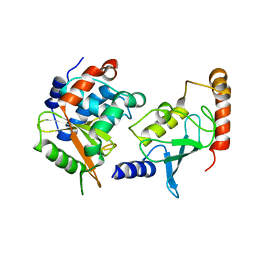 | | Complex structure of OspI and Ubc13 | | Descriptor: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | Deposit date: | 2013-01-09 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
177L
 
 | |
167L
 
 | |
