6NWP
 
 | | Chronic traumatic encephalopathy Type I Tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zivanov, J, Zhang, W, Murzin, A.G, Garringer, H.J, Vidal, R, Crowther, R.A, Newell, K.L, Ghetti, B, Goedert, M, Scheres, H.W. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules.
Nature, 568, 2019
|
|
6NWQ
 
 | | Chronic traumatic encephalopathy Type II Tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zivanov, J, Zhang, W, Murzin, A.G, Garringer, H.J, Vidal, R, Crowther, R.A, Newell, K.L, Ghetti, B, Goedert, M, Scheres, H.W. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules.
Nature, 568, 2019
|
|
5VBT
 
 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
2L84
 
 | | Solution NMR structures of CBP bromodomain with small molecule j28 | | Descriptor: | 5-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-2,4-dimethylbenzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
2PK9
 
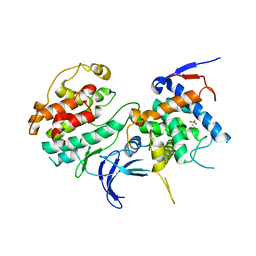 | | Structure of the Pho85-Pho80 CDK-cyclin Complex of the Phosphate-responsive Signal Transduction Pathway | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cyclin-dependent protein kinase PHO85, PHO85 cyclin PHO80 | | Authors: | Huang, K, Ferrin-O'Connell, I, Zhang, W, Leonard, G.A, O'Shea, E.K, Quiocho, F.A. | | Deposit date: | 2007-04-17 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of the Pho85-Pho80 CDK-Cyclin Complex of the Phosphate-Responsive Signal Transduction Pathway
Mol.Cell, 28, 2007
|
|
2PMI
 
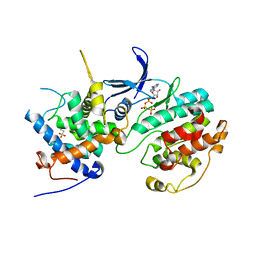 | | Structure of the Pho85-Pho80 CDK-cyclin Complex of the Phosphate-responsive Signal Transduction Pathway with Bound ATP-gamma-S | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cyclin-dependent protein kinase PHO85, PHO85 cyclin PHO80, ... | | Authors: | Huang, K, Ferrin-O'Connell, I, Zhang, W, Leonard, G.A, O'Shea, E.K, Quiocho, F.A. | | Deposit date: | 2007-04-23 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Pho85-Pho80 CDK-Cyclin Complex of the Phosphate-Responsive Signal Transduction Pathway
Mol.Cell, 28, 2007
|
|
5HCK
 
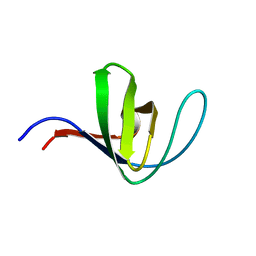 | | HUMAN HCK SH3 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
8HCZ
 
 | |
8HDF
 
 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
7DW5
 
 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | Descriptor: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | Authors: | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
7WA9
 
 | |
8ILB
 
 | | The complexes of RbcL, AtRaf1 and AtBSD2 (LFB) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8ILM
 
 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IO2
 
 | | The Rubisco assembly intermidate of Arabidopsis thaliana Rubisco accumulation factor 1 (AtRaf1) and Rubisco large subunit (RbcL) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1.2, chloroplastic | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IOJ
 
 | | The Rubisco assembly intermidiate of Rubisco large subunit (RbcL) and Arabidopsis thaliana Rubisco accumulation factor 1 (AtRaf1) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1.2, chloroplastic | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8IOL
 
 | | The complex of Rubisco large subunit (RbcL) | | Descriptor: | Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
3TFZ
 
 | | Crystal structure of Zhui aromatase/cyclase from Streptomcyes sp. R1128 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclase, POTASSIUM ION | | Authors: | Ames, B.D, Lee, M.Y, Moody, C, Zhang, W, Tang, Y, Wong, S.K, Tsai, S.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Characterization of ZhuI Aromatase/Cyclase from the R1128 Polyketide Pathway.
Biochemistry, 50, 2011
|
|
7VUL
 
 | |
8IQU
 
 | | Structure of MtbFadD23 with PhU-AMS | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Fatty-acid-CoA ligase FadD23 | | Authors: | Yan, M.R, Zhang, W. | | Deposit date: | 2023-03-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the development of potential inhibitors targeting FadD23 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
2HD3
 
 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
8JJM
 
 | |
7ESH
 
 | | Crystal structure of amylosucrase from Calidithermus timidus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | Authors: | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
7DEA
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
