7W3F
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W38
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.0_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3I
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SB_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3M
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD5_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3C
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3H
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W39
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3B
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED5_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3J
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3G
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
9IZN
 
 | | Crystal structure of HKU1A RBD bound to TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Wang, W, Xu, Y, Zhang, S. | | Deposit date: | 2024-08-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of human coronavirus HKU1 receptor binding domain bound to TMPRSS2 receptor
Hlife, 2024
|
|
7LUG
 
 | | Crystal structure of the pnRFP B30Y mutant | | Descriptor: | PHOSPHATE ION, Red Fluorescent pnRFP B30Y mutant | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
7LQO
 
 | | Crystal structure of a genetically encoded red fluorescent peroxynitrite biosensor, pnRFP | | Descriptor: | PHOSPHATE ION, red fluorescent peroxynitrite biosensor pnRFP | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-14 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
7OQ6
 
 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
5JYY
 
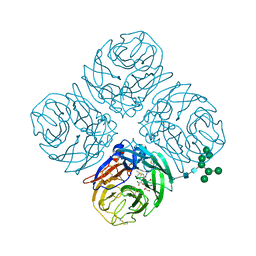 | | Structure-based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-resistant Influenza Viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-[(2-methoxyethyl)carbamoyl]-D-glycero-D-galacto-non-2-enon ic acid, CALCIUM ION, ... | | Authors: | Fu, L, Wu, Y, Bi, Y, Zhang, S, Lv, X, Qi, J, Li, Y, Lu, X, Yan, J, Gao, G.F, Li, X. | | Deposit date: | 2016-05-15 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-Resistant Influenza Viruses
J.Med.Chem., 59, 2016
|
|
9IJD
 
 | | Carazolol-activated human beta3 adrenergic receptor | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, Beta-3 adrenergic receptor, Camelid antibody VHH fragment, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular Mechanism of the beta 3AR Agonist Activity of a beta-Blocker.
Chempluschem, 2024
|
|
9IJE
 
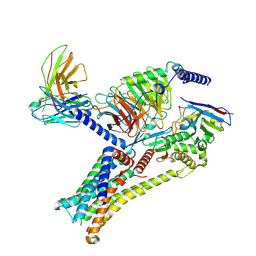 | | Epinephrine-activated human beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Camelid antibody VHH fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular Mechanism of the beta 3AR Agonist Activity of a beta-Blocker.
Chempluschem, 2024
|
|
5JMC
 
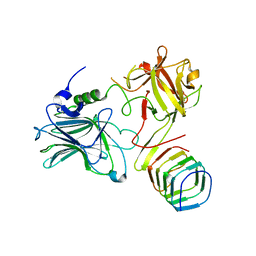 | | Receptor binding domain of Botulinum neurotoxin A in complex with rat SV2C | | Descriptor: | Botulinum neurotoxin type A, Synaptic vesicle glycoprotein 2C | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5IZ7
 
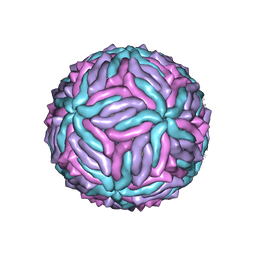 | | Cryo-EM structure of thermally stable Zika virus strain H/PF/2013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, structural protein E, structural protein M | | Authors: | Kostyuchenko, V.A, Zhang, S, Fibriansah, G, Lok, S.M. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the thermally stable Zika virus
Nature, 533, 2016
|
|
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
6QNS
 
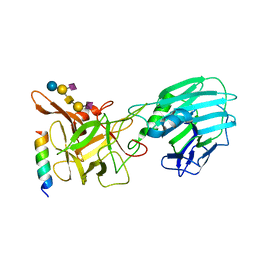 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B mutant I1248W/V1249W in complex with human synaptotagmin 1 and GD1a receptors | | Descriptor: | Botulinum neurotoxin type B, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Synaptotagmin-1 | | Authors: | Masuyer, G, Yin, L, Zhang, S, Miyashita, S.I, Dong, M, Stenmark, P. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy.
Plos Biol., 18, 2020
|
|
3LOI
 
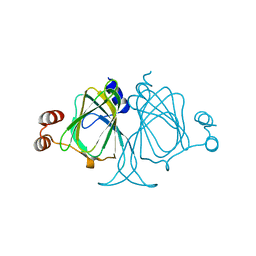 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in the apo and GDP-bound forms | | Descriptor: | Putative uncharacterized protein | | Authors: | Zou, C.Z, Du, Y, He, Y.-X, Saren, G, Zhang, X, Chen, Y, Zhang, S.-C. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
3LZZ
 
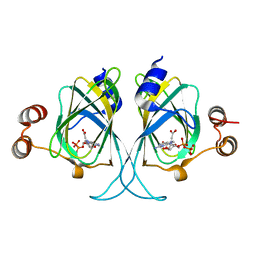 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in apo and GDP-bound forms | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | Authors: | Du, Y, He, Y.-X, Saren, G, Zhang, X, Zhang, S.-C, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-03-02 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
