8WR4
 
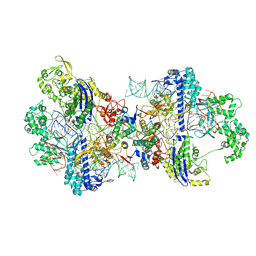 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
5XYN
 
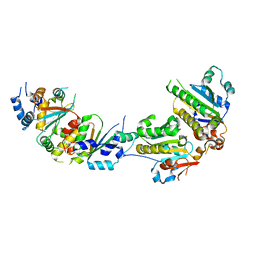 | | The crystal structure of Csm2-Psy3-Shu1-Shu2 complex from budding yeast | | Descriptor: | Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3, Suppressor of HU sensitivity involved in recombination protein 1, ... | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2017-07-09 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the functional role of the Shu complex in homologous recombination.
Nucleic Acids Res., 45, 2017
|
|
6D02
 
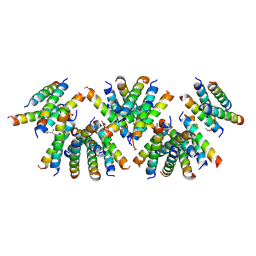 | | Cross-alpha Amyloid-like Structure alphaAmL, 2nd form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.-Q, Liu, L, Degrado, W.F. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
8WMW
 
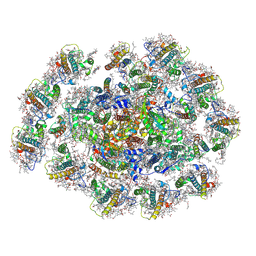 | | The structure of PSI-11CAC at the stationary growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WMM
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WWU
 
 | | 1-naphthylamine GS in complex with AMP PNP | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
8WMH
 
 | | Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMN
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WNW
 
 | | the structure of PsaQ | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, PsaQ | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
7BXU
 
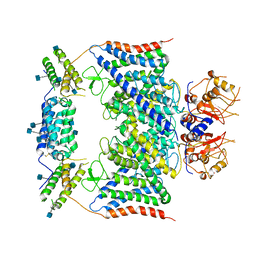 | | CLC-7/Ostm1 membrane protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insights into the human CLC-7/Ostm1 transporter.
Sci Adv, 6, 2020
|
|
5G04
 
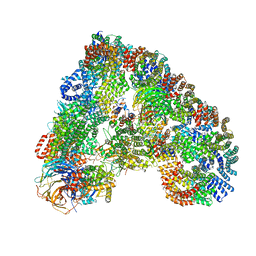 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5G05
 
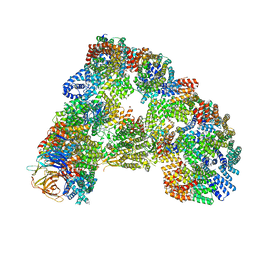 | | Cryo-EM structure of combined apo phosphorylated APC | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
7YPG
 
 | |
7N6W
 
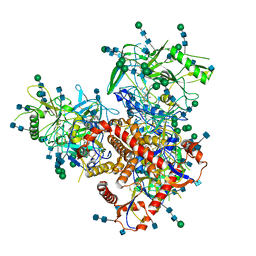 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
7N6U
 
 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
5ZHH
 
 | |
5GN8
 
 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
5WLL
 
 | |
5WLJ
 
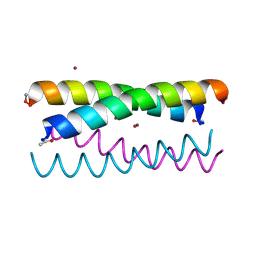 | |
5WLM
 
 | |
7BUA
 
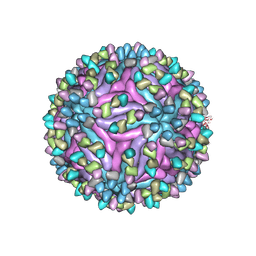 | | Cryo-EM structure of zika virus complexed with Fab SIgN-3C at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, SIgN-3C Fab heavy chain, ... | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
7BUD
 
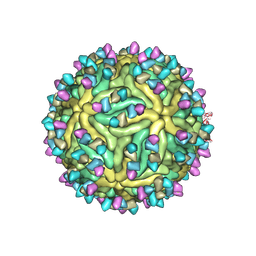 | | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dengue virus serotype 2 E protein, ... | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
7BUE
 
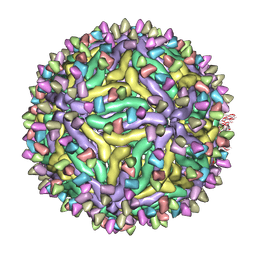 | | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 5.0 | | Descriptor: | Dengue serotype 2 E protein ectodomain, SIgN-3C Fab heavy chain, SIgN-3C Fab light chain | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
7BU8
 
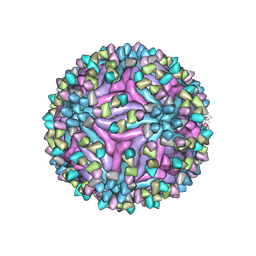 | | Cryo-EM structure of zika virus complexed with Fab SIgN-3C at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, SIgN-3C Fab heavy chain, ... | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-05 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
7BUB
 
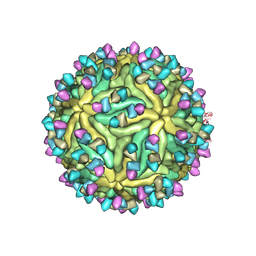 | | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dengue virus serotype 2 M protein, ... | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
