3N7Z
 
 | | Crystal structure of acetyltransferase from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, SODIUM ION | | Authors: | Chang, C, Wu, R, Gornicki, P, Zhang, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and Structural Analysis of an Eis Family Aminoglycoside Acetyltransferase from Bacillus anthracis.
Biochemistry, 54, 2015
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
6UV7
 
 | |
6UVI
 
 | |
8Q20
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425D mutant (A. marina) | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vanadium-dependent bromoperoxidase, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Unraveling the Molecular Basis of Substrate Specificity and Halogen Activation in Vanadium-Dependent Haloperoxidases
to be published
|
|
8Q21
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant (A. marina) | | Descriptor: | PHOSPHATE ION, SODIUM ION, Vanadium-dependent bromoperoxidase, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the Molecular Basis of Substrate Specificity and Halogen Activation in Vanadium-Dependent Haloperoxidases
to be published
|
|
8Q22
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant in complex with 1,3,5-trimethoxybenzene (A. marina) | | Descriptor: | 1,3,5-trimethoxybenzene, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the Molecular Basis of Substrate Specificity and Halogen Activation in Vanadium-Dependent Haloperoxidases
to be published
|
|
8OTZ
 
 | | 48-nm repeat of the native axonemal doublet microtubule from bovine sperm | | Descriptor: | ATP6V1F neighbor, CFAP97 domain containing 1, Chromosome 13 C20orf85 homolog, ... | | Authors: | Leung, M.R, Zeng, J, Zhang, R, Zeev-Ben-Mordehai, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural specializations of the sperm tail.
Cell, 186, 2023
|
|
8ZDZ
 
 | |
8ZE3
 
 | |
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
3CQV
 
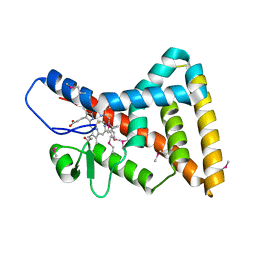 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
7W0W
 
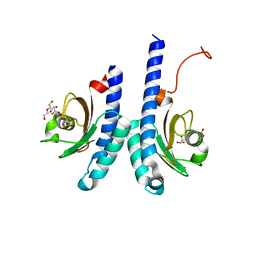 | | The novel membrane-proximal sensing mechanism in a broad-ligand binding chemoreceptor McpA of Bacillus velezensis | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Feng, H.C, Shen, Q.R, Zhang, R.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Signal binding at both modules of its dCache domain enables the McpA chemoreceptor of Bacillus velezensis to sense different ligands.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
3D6K
 
 | | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Tan, K, Zhang, R, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae
To be Published
|
|
8ZE0
 
 | |
8ZE2
 
 | |
2GAU
 
 | | Crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis (APC80792), Structural genomics, MCSG | | Descriptor: | transcriptional regulator, Crp/Fnr family | | Authors: | Rotella, F.J, Zhang, R.G, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis
To be Published
|
|
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
2HMC
 
 | | The Crystal Structure of Dihydrodipicolinate Synthase DapA from Agrobacterium tumefaciens | | Descriptor: | Dihydrodipicolinate synthase, MAGNESIUM ION | | Authors: | Kim, Y, Zhang, R, Xu, X, Zheng, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Dihydrodipicolinate Synthase DapA from Agrobacterium tumefaciens
To be Published, 2006
|
|
2OQW
 
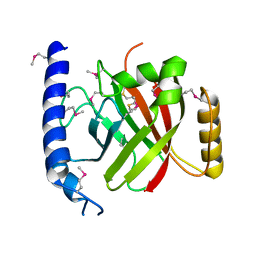 | | The crystal structure of sortase B from B.anthracis in complex with AAEK1 | | Descriptor: | Sortase B | | Authors: | Wu, R, Zhang, R, Marresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2OQZ
 
 | | The crystal structure of sortase B from B.anthracis in complex with AAEK2 | | Descriptor: | ACETIC ACID, Sortase B | | Authors: | Wu, R, Zhang, R, Maresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2I5U
 
 | | Crystal structure of DnaD domain protein from Enterococcus faecalis. Structural genomics target APC85179 | | Descriptor: | DnaD domain protein, MAGNESIUM ION | | Authors: | Wu, R, Zhang, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 A crystal structure of a DnaD domain protein from Enterococcus Faecalis
To be Published, 2006
|
|
