4V8M
 
 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | Descriptor: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | Authors: | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | Deposit date: | 2012-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.57 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
1HQB
 
 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G, Neuhaus, F.C. | | Deposit date: | 2000-12-14 | | Release date: | 2001-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
1F95
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | Descriptor: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | Authors: | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
1F96
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND NNOS PEPTIDE COMPLEX | | Descriptor: | DYNEIN LIGHT CHAIN 8, PROTEIN (NNOS, NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Fan, J.S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
1F3C
 
 | |
1XWN
 
 | | solution structure of cyclophilin like 1(PPIL1) and insights into its interaction with SKIP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase like 1 | | Authors: | Xu, C, Xu, Y, Tang, Y, Wu, J, Shi, Y, Huang, Q, Zhang, Q. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human peptidyl prolyl isomerase like protein 1 and insights into its interaction with SKIP
J.Biol.Chem., 281, 2006
|
|
4Y33
 
 | | Crystal of NO66 in complex with Ni(II)and N-oxalylglycine (NOG) | | Descriptor: | Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5IWW
 
 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZI
 
 | | Crystal Structure of TR3 LBD in complex with DPDO | | Descriptor: | 1-(3,5-dimethoxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
1DV5
 
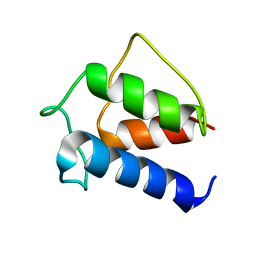 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G.C, Neuhaus, F.C. | | Deposit date: | 2000-01-19 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
2QCO
 
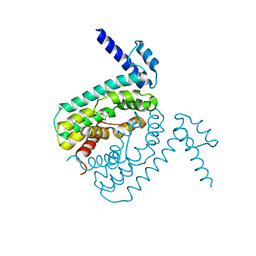 | | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni | | Descriptor: | CmeR, GLYCEROL | | Authors: | Gu, R, Su, C, Shi, F, Li, M, McDermott, G, Zhang, Q, Yu, E.W. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni.
J.Mol.Biol., 372, 2007
|
|
3HTO
 
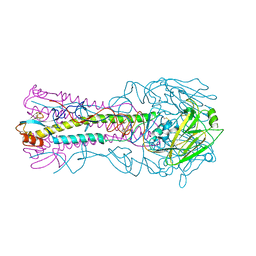 | | the hemagglutinin structure of an avian H1N1 influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
3HTP
 
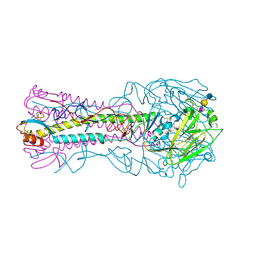 | | the hemagglutinin structure of an avian H1N1 influenza A virus in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
3HTQ
 
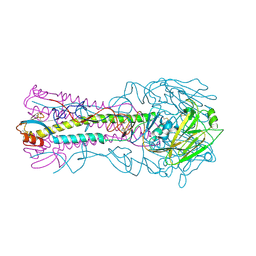 | | the hemagglutinin structure of an avian H1N1 influenza A virus in complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
3HTT
 
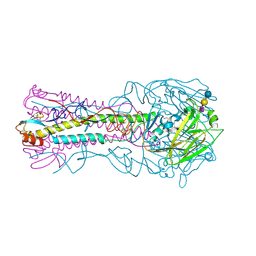 | | The hemagglutinin structure of an avian H1N1 influenza A virus in complex with 2,3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
5X0R
 
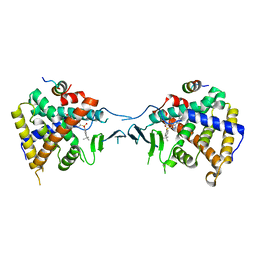 | | Crystal Structure of PXR LBD Complexed with SJB7 | | Descriptor: | 4-[(4-tert-butylphenyl)sulfonyl]-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Lv, L, Lin, W, Chai, S.C, Zhang, Q, Chen, T. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | SPA70 is a potent antagonist of human pregnane X receptor.
Nat Commun, 8, 2017
|
|
5WCH
 
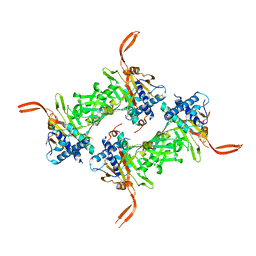 | | Crystal structure of the catalytic domain of human USP9X | | Descriptor: | Probable ubiquitin carboxyl-terminal hydrolase FAF-X, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2RNW
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNY
 
 | | Complex Structures of CBP Bromodomain with H4 ack20 Peptide | | Descriptor: | CREB-binding protein, Histone H4 | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2W80
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
2W81
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
