5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
8K5Q
 
 | | Crystal structure of YajQ STM0435 with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), YajQ | | Authors: | Dai, Y, Zhang, M, Wang, W, Li, B. | | Deposit date: | 2023-07-23 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A c-di-GMP binding effector STM0435 modulates flagellar motility and pathogenicity in Salmonella
Virulence, 15, 2024
|
|
8K4I
 
 | |
6JLE
 
 | | Crystal structure of MORN4/Myo3a complex | | Descriptor: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | Authors: | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
6JVX
 
 | | Crystal structure of RBM38 in complex with RNA | | Descriptor: | RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
6A9X
 
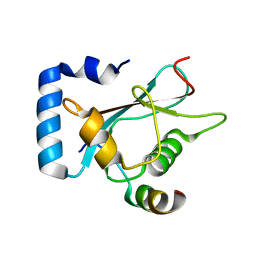 | | Crystal Structure of AnkG/GABARAP Complex | | Descriptor: | Ankyrin-3, Gamma-aminobutyric acid receptor-associated protein | | Authors: | Wang, C, Li, J, Chen, K, Zhang, M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Ankyrin-G regulates forebrain connectivity and network synchronization via interaction with GABARAP.
Mol. Psychiatry, 2018
|
|
4D8O
 
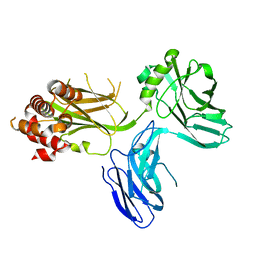 | |
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6JVY
 
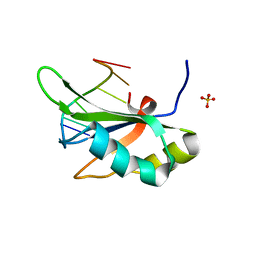 | | Crystal structure of RBM38 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*GP*TP*GP*TP*GP*TP*GP*TP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
6JEA
 
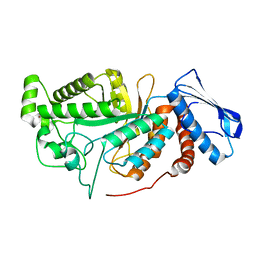 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6KY4
 
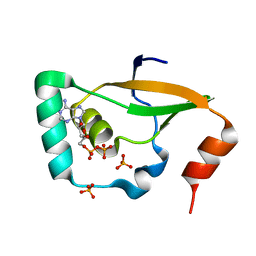 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6JE8
 
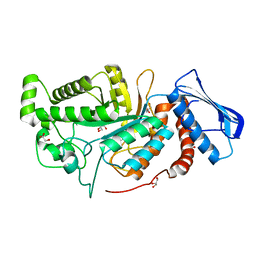 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | Beta-N-acetylhexosaminidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6IRR
 
 | | Solution structure of DISC1/ATF4 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Cyclic AMP-dependent transcription factor ATF-4 | | Authors: | Ye, F, Yu, C, Zhang, M. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Mol. Psychiatry, 2019
|
|
7YFF
 
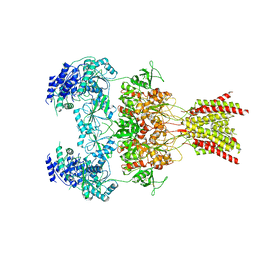 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
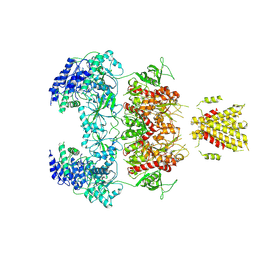 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFR
 
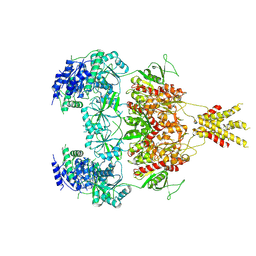 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XS7
 
 | | structure of a membrane-integrated glycosyltransferase | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, Chitin synthase 1, DODECANE, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
7XS6
 
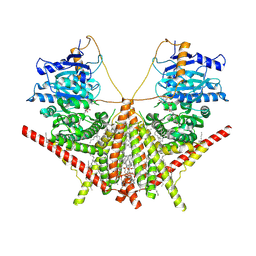 | | structure of a membrane-integrated glycosyltransferase with inhibitor | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
2LSR
 
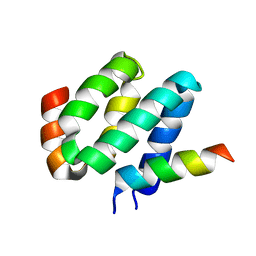 | | Solution structure of harmonin N terminal domain in complex with a exon68 encoded peptide of cadherin23 | | Descriptor: | Harmonin, peptide from Cadherin-23 | | Authors: | Pan, L, Wu, L, Zhang, C, Zhang, M. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia.
J.Biol.Chem., 287, 2012
|
|
5Y1Z
 
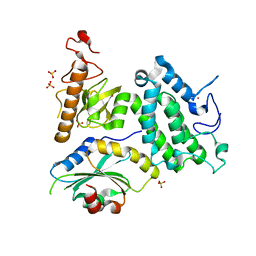 | | Crystal structure of ZMYND8 PHD-BROMO-PWWP tandem in complex with Drebrin ADF-H domain | | Descriptor: | Drebrin, GLYCEROL, Protein kinase C-binding protein 1, ... | | Authors: | Yao, N, Li, J, Liu, H, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2017-07-22 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | The Structure of the ZMYND8/Drebrin Complex Suggests a Cytoplasmic Sequestering Mechanism of ZMYND8 by Drebrin
Structure, 25, 2017
|
|
6JEB
 
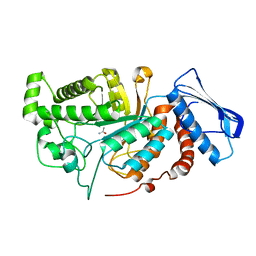 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | ACETAMIDE, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5YZY
 
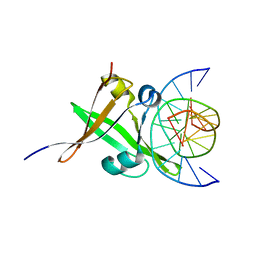 | | AtVAL1 B3 domain in complex with 13bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*A)-3'), ... | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
5ZTE
 
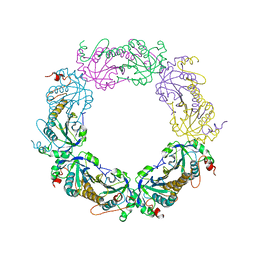 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | Descriptor: | 2-Cys peroxiredoxin BAS1, chloroplastic | | Authors: | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5Z00
 
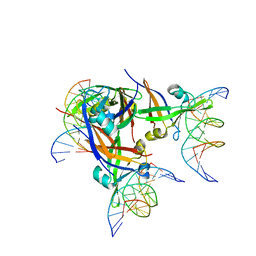 | | AtVAL1 B3 domain in complex with 15bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), ... | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
5YZZ
 
 | | AtVAL1 B3 domain in complex with 13bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*A)-3') | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
