5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
1H74
 
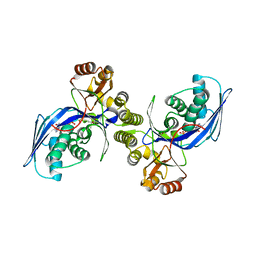 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH ILE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOMOSERINE KINASE, ISOLEUCINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
4YVP
 
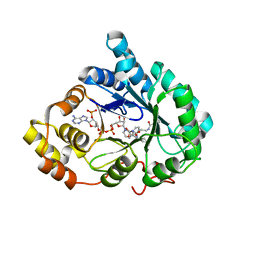 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
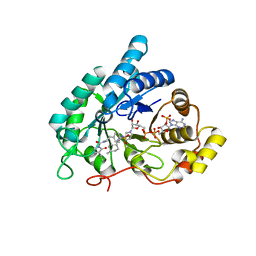 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVV
 
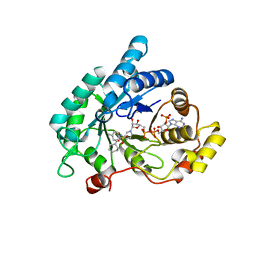 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
4KYK
 
 | | Crystal structure of mouse glyoxalase I complexed with indomethacin | | Descriptor: | INDOMETHACIN, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
6OMV
 
 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4-DNA complex | | Descriptor: | AcrVA4, Cpf1, DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*GP*GP*A)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
4JIH
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Epalrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-[(2E)-2-methyl-3-phenylprop-2-en-1-ylidene]-4-oxo-2-thioxo-1,3-thiazolidin-3-yl}acetic acid | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JIR
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Epalrestat | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X. | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
1XMA
 
 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
4JII
 
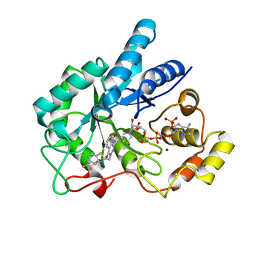 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1GVP
 
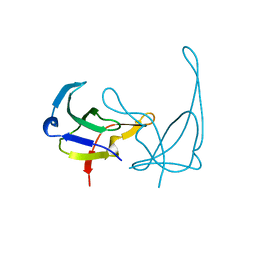 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1QKK
 
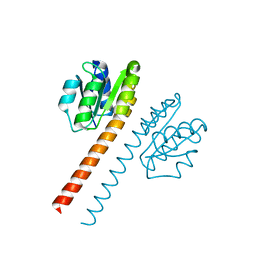 | | Crystal structure of the receiver domain and linker region of DctD from Sinorhizobium meliloti | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Meyer, M.G, Park, S, Zeringue, L, Staley, M, Mckinstry, M, Kaufman, R.I, Zhang, H, Yan, D, Yennawar, N, Farber, G.K, Nixon, B.T. | | Deposit date: | 1999-07-23 | | Release date: | 2000-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD.
Faseb J., 15, 2001
|
|
6LB3
 
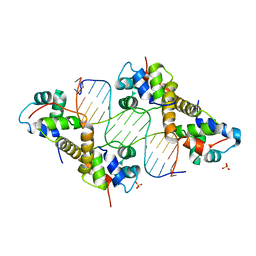 | | Crystal structure of PA4674 in complex with its operator DNA (18bp) from Pseudomonas aeruginosa | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*TP*TP*AP*AP*CP*GP*TP*TP*AP*AP*GP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*CP*TP*TP*AP*AP*CP*GP*TP*TP*AP*AP*GP*GP*GP*T)-3'), HTH cro/C1-type domain-containing protein, ... | | Authors: | Liu, Y, Zhang, H, Gao, Z, Dong, Y. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structure of PA4674 in complex with its operator DNA (18bp) from Pseudomonas aeruginosa
To Be Published
|
|
6LW5
 
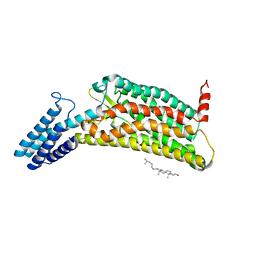 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
3R1A
 
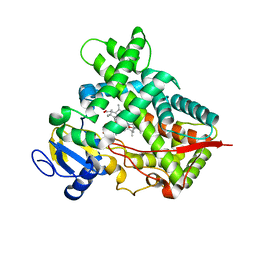 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3R1B
 
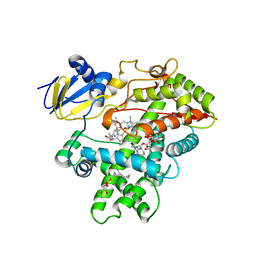 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
1LW7
 
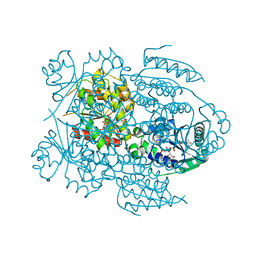 | | NADR PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, TRANSCRIPTIONAL REGULATOR NADR | | Authors: | Singh, S.K, Kurnasov, O.V, Chen, B, Robinson, H, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-05-30 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities.
J.Biol.Chem., 277, 2002
|
|
1RFS
 
 | | RIESKE SOLUBLE FRAGMENT FROM SPINACH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE PROTEIN | | Authors: | Carrell, C.J, Zhang, H, Cramer, W.A, Smith, J.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein.
Structure, 5, 1997
|
|
1XRG
 
 | | Conserved hypothetical protein from Clostridium thermocellum Cth-2968 | | Descriptor: | Putative translation initiation inhibitor, yjgF family, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Zhang, H, Arendall III, W.B, Ljundahl, L, Liu, Z.-J, Rose, J, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-14 | | Release date: | 2004-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved hypothetical protein from Clostridium thermocellum Cth-2968
To be published
|
|
