8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
1RQL
 
 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1RQN
 
 | | Phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1LVH
 
 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
1O03
 
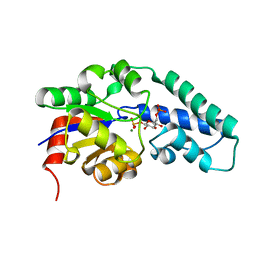 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 6-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
1O08
 
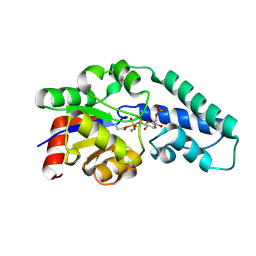 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 1-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
1P4X
 
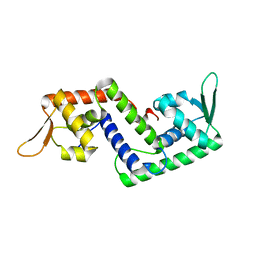 | | Crystal structure of SarS protein from Staphylococcus Aureus | | Descriptor: | staphylococcal accessory regulator A homologue | | Authors: | Li, R, Manna, A.C, Dai, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SarS protein from Staphylococcus aureus
J.BACTERIOL., 185, 2003
|
|
2HFR
 
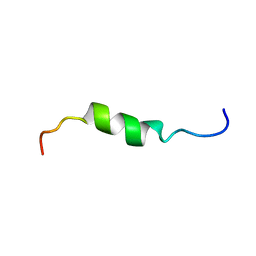 | | solution structure of antimicrobial peptide Fowlicidin 3 | | Descriptor: | Fowlicidin-3 | | Authors: | Bommineni, Y.R, Dai, H, Gong, Y, Prakash, O, Zhang, G. | | Deposit date: | 2006-06-26 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities.
Febs J., 274, 2007
|
|
1XDY
 
 | | Structural and Biochemical Identification of a Novel Bacterial Oxidoreductase, W-containing cofactor | | Descriptor: | Bacterial Sulfite Oxidase, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, TUNGSTEN ION | | Authors: | Loschi, L, Brokx, S.J, Hills, T.L, Zhang, G, Bertero, M.G, Lovering, A.L, Weiner, J.H, Strynadka, N.C. | | Deposit date: | 2004-09-08 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical identification of a novel bacterial oxidoreductase.
J.Biol.Chem., 279, 2004
|
|
1XDQ
 
 | | Structural and Biochemical Identification of a Novel Bacterial Oxidoreductase | | Descriptor: | Bacterial Sulfite Oxidase, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Loschi, L, Brokx, S.J, Hills, T.L, Zhang, G, Bertero, M.G, Lovering, A.L, Weiner, J.H, Strynadka, N.C. | | Deposit date: | 2004-09-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical identification of a novel bacterial oxidoreductase.
J.Biol.Chem., 279, 2004
|
|
1Z4N
 
 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate cocrystallized with Fluoride | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
1Z4O
 
 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
2LQV
 
 | | YebF | | Descriptor: | Protein yebF | | Authors: | Prehna, G, Zhang, G, Gong, X, Duszyk, M, Okon, M, Mcintosh, L.P, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2012-03-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Protein Export Pathway Involving Escherichia coli Porins.
Structure, 20, 2012
|
|
1JH5
 
 | | Crystal Structure of sTALL-1 of TNF family ligand | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 13B | | Authors: | Liu, Y, Xu, L, Opalka, N, Shu, H.-B, Zhang, G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands.
Cell(Cambridge,Mass.), 108, 2002
|
|
1RDF
 
 | | G50P mutant of phosphonoacetaldehyde hydrolase in complex with substrate analogue vinyl sulfonate | | Descriptor: | ETHANESULFONIC ACID, MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-11-05 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the substrate specificity loop of the HAD superfamily cap domain
Biochemistry, 43, 2004
|
|
3LD8
 
 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LDB
 
 | | Structure of JMJD6 complexd with ALPHA-KETOGLUTARATE and Fab Fragment. | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2IOF
 
 | | Crystal structure of phosphonoacetaldehyde hydrolase with sodium borohydride-reduced substrate intermediate | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphonoacetaldehyde hydrolase | | Authors: | Allen, K.A, Lahiri, S.D, Zhang, G, Dunaway-Mariano, D. | | Deposit date: | 2006-10-10 | | Release date: | 2007-07-17 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage.
Bioorg.Chem., 34, 2006
|
|
2IOH
 
 | | Crystal structure of phosphonoacetaldehyde hydrolase with a K53R mutation | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphonoacetaldehyde hydrolase | | Authors: | Allen, K.A, Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Peisach, E. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage.
Bioorg.Chem., 34, 2006
|
|
5JFB
 
 | | Crystal structure of the scavenger receptor cysteine-rich domain 5 (SRCR5) from porcine CD163 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Jiang, L, Qiao, S, Zhang, G, Li, R. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection
J. Virol., 91, 2017
|
|
2GP3
 
 | | Crystal structure of the catalytic core domain of jmjd2a | | Descriptor: | FE (II) ION, Jumonji domain-containing protein 2A, ZINC ION | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
2GP5
 
 | | Crystal structure of catalytic core domain of jmjd2A complexed with alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Jumonji domain-containing protein 2A, ... | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
2JZC
 
 | | NMR solution structure of ALG13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Northeast Structural Genomics Consortium target YG1 | | Descriptor: | UDP-N-acetylglucosamine transferase subunit ALG13 | | Authors: | Wang, X, Weldeghorghis, T, Zhang, G, Imepriali, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase.
Structure, 16, 2008
|
|
