1SWV
 
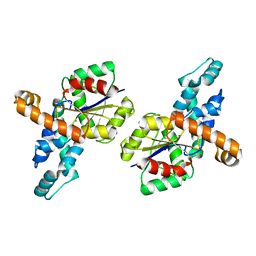 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1SWW
 
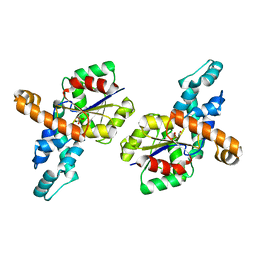 | | Crystal structure of the phosphonoacetaldehyde hydrolase D12A mutant complexed with magnesium and substrate phosphonoacetaldehyde | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1OQD
 
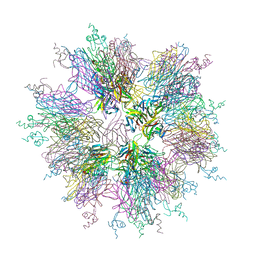 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQE
 
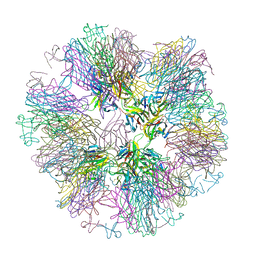 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1HSJ
 
 | | SARR MBP FUSION STRUCTURE | | Descriptor: | FUSION PROTEIN CONSISTING OF STAPHYLOCOCCUS ACCESSORY REGULATOR PROTEIN R AND MALTOSE BINDING PROTEIN, alpha-D-glucopyranose | | Authors: | Zhang, G. | | Deposit date: | 2000-12-26 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the SarR protein from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1PTQ
 
 | | PROTEIN KINASE C DELTA CYS2 DOMAIN | | Descriptor: | PROTEIN KINASE C DELTA TYPE, ZINC ION | | Authors: | Zhang, G, Hurley, J.H. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester.
Cell(Cambridge,Mass.), 81, 1995
|
|
1PTR
 
 | |
1ZOL
 
 | | native beta-PGM | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Zhang, G, Tremblay, L.W, Dai, J, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-05-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic cycling in beta-phosphoglucomutase: a kinetic and structural analysis
Biochemistry, 44, 2005
|
|
2LSP
 
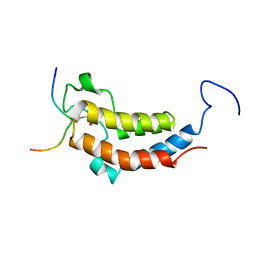 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
1AB8
 
 | |
1BDF
 
 | |
2P5B
 
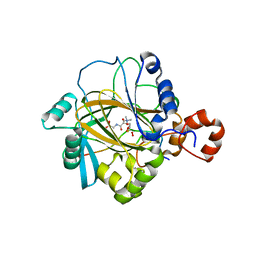 | | The complex structure of JMJD2A and trimethylated H3K36 peptide | | Descriptor: | FE (II) ION, Histone H3, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Zhang, G, Chen, Z, Zang, J, Hong, X, Shi, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2007-06-12 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7F0X
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F12
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F15
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
5HRJ
 
 | |
1O03
 
 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 6-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
2W7X
 
 | | Cellular inhibition of checkpoint kinase 2 and potentiation of cytotoxic drugs by novel Chk2 inhibitor PV1019 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[4-[(E)-N-carbamimidamido-C-methyl-carbonimidoyl]phenyl]-7-nitro-1H-indole-2-carboxamide, ... | | Authors: | Jobson, A.G, Lountos, G.T, Lorenzi, P.L, Llamas, J, Connelly, J, Tropea, J.E, Onda, A, Kondapaka, S, Zhang, G, Caplen, N.J, Caredellina, J.H, Monks, A, Self, C, Waugh, D.S, Shoemaker, R.H, Pommier, Y. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cellular Inhibition of Chk2 Kinase and Potentiation of Camptothecins and Radiation by the Novel Chk2 Inhibitor Pv1019.
J.Pharmacol.Exp.Ther., 331, 2009
|
|
1FEZ
 
 | | THE CRYSTAL STRUCTURE OF BACILLUS CEREUS PHOSPHONOACETALDEHYDE HYDROLASE COMPLEXED WITH TUNGSTATE, A PRODUCT ANALOG | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE HYDROLASE, TUNGSTATE(VI)ION | | Authors: | Morais, M.C, Zhang, W, Baker, A.S, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2000-07-24 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily.
Biochemistry, 39, 2000
|
|
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
1RQN
 
 | | Phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1RQL
 
 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1LVH
 
 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
2AMN
 
 | | Solution structure of Fowlicidin-1, a novel Cathelicidin antimicrobial peptide from chicken | | Descriptor: | cathelicidin | | Authors: | Xiao, Y, Dai, H, Bommineni, Y.R, Prakash, O, Zhang, G. | | Deposit date: | 2005-08-09 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken.
Febs J., 273, 2006
|
|
1O08
 
 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 1-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
