5Z9Y
 
 | | Crystal structure of Mycobacterium tuberculosis thiazole synthase (ThiG) complexed with DXP | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Thiazole synthase | | Authors: | Zhang, J, Zhang, B, Zhao, Y, Yang, X, Huang, M, Cui, P, Zhang, W, Li, J, Zhang, Y. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Snapshots of catalysis: Structure of covalently bound substrate trapped in Mycobacterium tuberculosis thiazole synthase (ThiG).
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5K6G
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-24 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6B
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9 DS-Cav1 variant. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Rundlet, E.J, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6F
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-19 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0 | | Authors: | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-10 | | Last modified: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6C
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-12 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (3.576 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6H
 
 | |
5K6I
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 A149C-Y458C, S46G-E92D-S215P-K465Q variant. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1L3Q
 
 | | H. rufescens abalone shell Lustrin A consensus repeat, FPGKNVNCTSGE, pH 7.4, 1-H NMR structure | | Descriptor: | Lustrin A | | Authors: | Evans, J.S, Wustman, B.A, Zhang, B, Morse, D.E. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Model peptide studies of sequence regions in the elastomeric biomineralization protein, Lustrin A. I. The C-domain consensus-PG-, -NVNCT-motif
Biopolymers, 63, 2002
|
|
4R7A
 
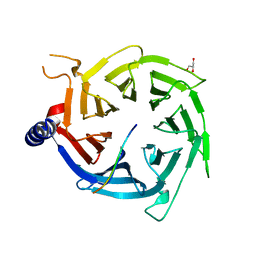 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
8HXQ
 
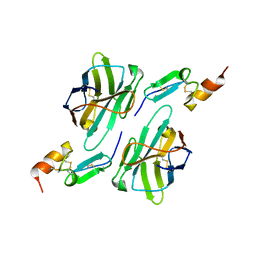 | | Nanobody1 in complex with human BCMA ECD | | Descriptor: | Nanobody1, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Sun, Y, Zhang, B. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
8HXR
 
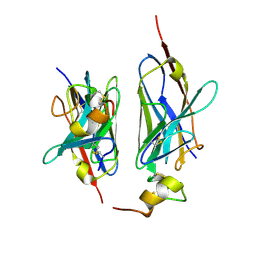 | | Nanobody2 in complex with human BCMA ECD | | Descriptor: | Nanobody2, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Sun, Y, Zhang, B. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
5YGE
 
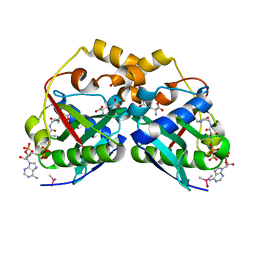 | | ArgA complexed with AceCoA and glutamate | | Descriptor: | ACETYL COENZYME *A, Amino-acid acetyltransferase, CACODYLIC ACID, ... | | Authors: | Yang, X, Wu, L, Ran, Y, Xu, A, Zhang, B, Yang, X, Zhang, R, Rao, Z, Li, J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Crystal structure of l-glutamate N-acetyltransferase ArgA from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1865, 2017
|
|
3I9N
 
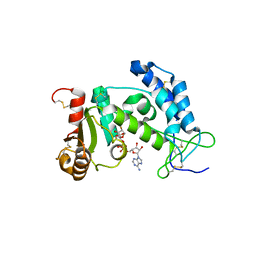 | | Crystal structure of human CD38 complexed with an analog ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9K
 
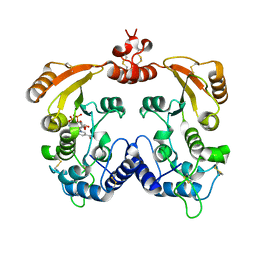 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9M
 
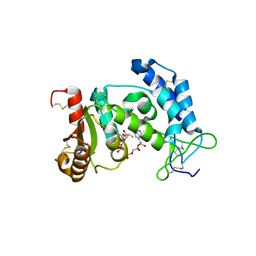 | | Crystal structure of human CD38 complexed with an analog ara-2'F-ADPR | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9J
 
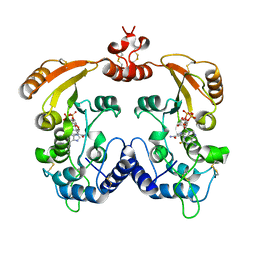 | | Crystal structure of ADP ribosyl cyclase complexed with a substrate analog and a product nicotinamide | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE, Nicotinamide 2-fluoro-adenine dinucleotide, ... | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9L
 
 | | Crystal structure of ADP ribosyl cyclase complexed with N1-cIDPR | | Descriptor: | ADP-ribosyl cyclase, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
6DF6
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16ab | | Descriptor: | (8R)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, GLYCEROL | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5LUS
 
 | | Structures of DHBN domain of Pelecanus crispus BLM helicase | | Descriptor: | BLM helicase | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5MK5
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | Bloom syndrome protein, IODIDE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LUT
 
 | | Structures of DHBN domain of Gallus gallus BLM helicase | | Descriptor: | BLM helicase, PHOSPHATE ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LUP
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | BLM protein, PHOSPHATE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
8XB9
 
 | | The Crystal Structure of polo-box domain of PLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase PLK1 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Meng, Q, Zhang, B. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of polo-box domain of PLK1 from Biortus
To Be Published
|
|
5Y20
 
 | |
5YC4
 
 | |
