3NMZ
 
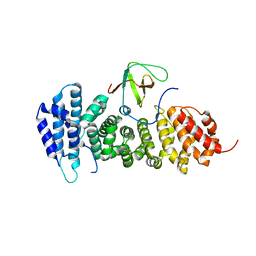 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3NMW
 
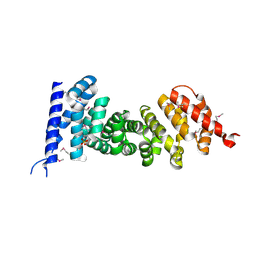 | | Crystal structure of armadillo repeats domain of APC | | Descriptor: | APC variant protein, SULFATE ION | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3JAB
 
 | | Domain organization and conformational plasticity of the G protein effector, PDE6 | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-05-26 | | Release date: | 2015-06-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
3NMX
 
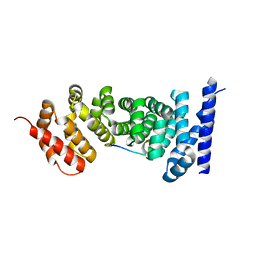 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3H6E
 
 | |
3GV1
 
 | |
3D8V
 
 | |
1BG5
 
 | | CRYSTAL STRUCTURE OF THE ANKYRIN BINDING DOMAIN OF ALPHA-NA,K-ATPASE AS A FUSION PROTEIN WITH GLUTATHIONE S-TRANSFERASE | | Descriptor: | FUSION PROTEIN OF ALPHA-NA,K-ATPASE WITH GLUTATHIONE S-TRANSFERASE | | Authors: | Zhang, Z, Devarajan, P, Morrow, J.S. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the ankyrin-binding domain of alpha-Na,K-ATPase.
J.Biol.Chem., 273, 1998
|
|
3SSZ
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium
To be Published
|
|
3DA8
 
 | | Crystal structure of PurN from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-05-28 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3DCJ
 
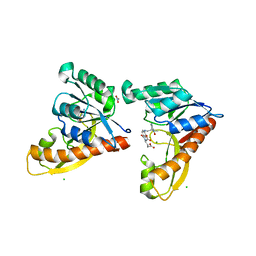 | | Crystal structure of glycinamide formyltransferase (PurN) from Mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-tetrahydrofolic acid derivative | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-06-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3VA9
 
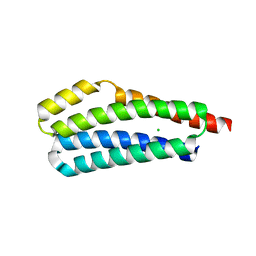 | |
7E7X
 
 | |
7YTP
 
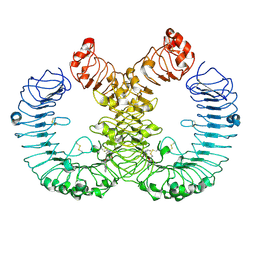 | | TLR7 in complex with an inhibitor | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2022-08-15 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | TLR7 in complex with an inhibitor
To Be Published
|
|
8HD0
 
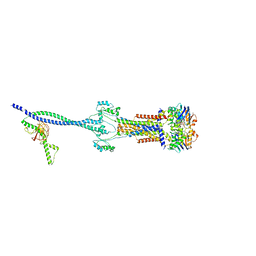 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Chen, Y. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insight into the septal peptidoglycan hydrolysis machinery of bacterial cell division
To Be Published
|
|
3V2H
 
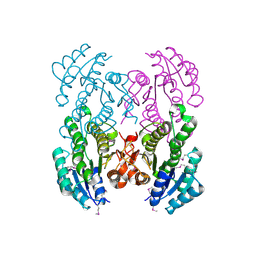 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3RL8
 
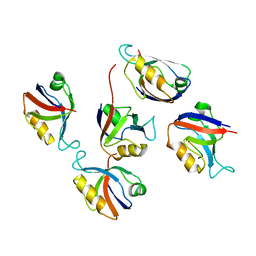 | | Crystal structure of hDLG1-PDZ2 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
6BRR
 
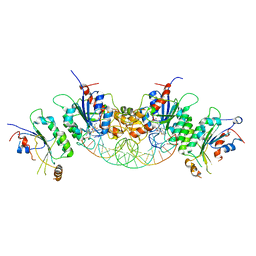 | |
7FH5
 
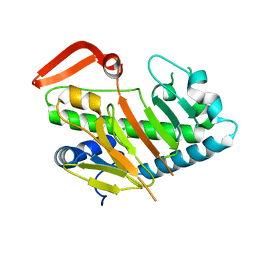 | | Structure of AdaV | | Descriptor: | AdaV, CHLORIDE ION, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
4Z4P
 
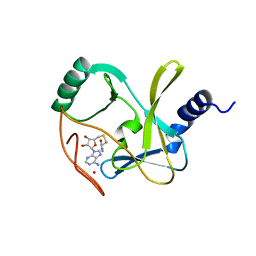 | | Structure of the MLL4 SET Domain | | Descriptor: | Histone-lysine N-methyltransferase 2D, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Zhang, Z, Mittal, A, Reid, J, Reich, S, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolving Catalytic Properties of the MLL Family SET Domain.
Structure, 23, 2015
|
|
3OVG
 
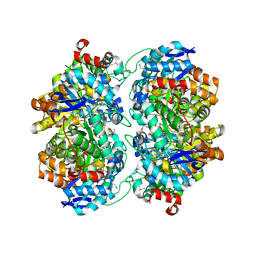 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
3ZN3
 
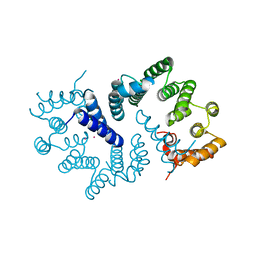 | | N-terminal domain of S. pombe Cdc23 APC subunit | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8, MERCURY (II) ION | | Authors: | Zhang, Z, Yang, J, Conin, N, Kulkarni, K, Barford, D. | | Deposit date: | 2013-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Four Canonical Tpr Subunits of Human Apc/C Form Related Homo-Dimeric Structures and Stack in Parallel to Form a Tpr Suprahelix
J.Mol.Biol., 425, 2013
|
|
5ZSL
 
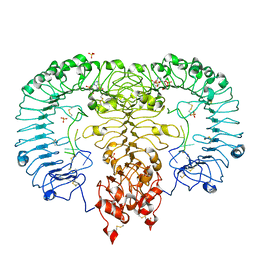 | | Crystal structure of monkey TLR7 in complex with GGUUGG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2S,3aR,4R,6R,6aR)-2-hydroxy-6-(hydroxymethyl)-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]-3,9-dihydro-6H-purin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSH
 
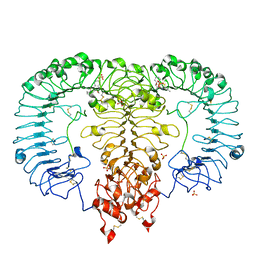 | | Crystal structure of monkey TLR7 in complex with CL075 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-propyl[1,3]thiazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSD
 
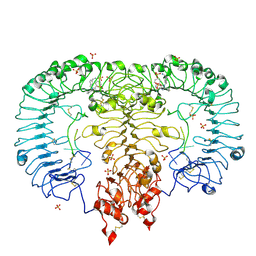 | | Crystal structure of monkey TLR7 in complex with IMDQ and GGUUGG | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
