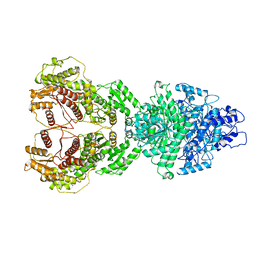
8HI4
 
 | |
5XBM
 
 | | Structure of SCARB2-JL2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | 著者 | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | 登録日 | 2017-03-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
4O67
 
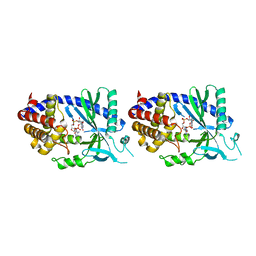 | | Human cyclic GMP-AMP synthase (cGAS) in complex with GAMP | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.444 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O68
 
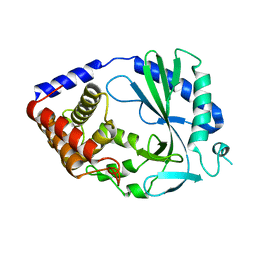 | | Structure of human cyclic GMP-AMP synthase (cGAS) | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.436 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O69
 
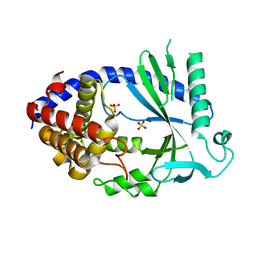 | | Human cyclic GMP-AMP synthase (cGAS) in complex with sulfate ion | | 分子名称: | Cyclic GMP-AMP synthase, SULFATE ION, ZINC ION | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O6A
 
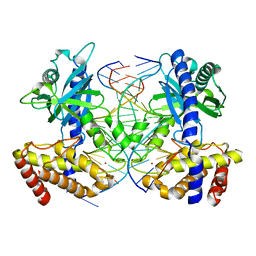 | | Mouse cyclic GMP-AMP synthase (cGAS) in complex with DNA | | 分子名称: | Cyclic GMP-AMP synthase, DNA1, DNA2, ... | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.859 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
3KL1
 
 | |
3KLX
 
 | |
3F4M
 
 | | Crystal structure of TIPE2 | | 分子名称: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | 著者 | Zhang, X, Wang, J, Shi, Y. | | 登録日 | 2008-11-01 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
5K3H
 
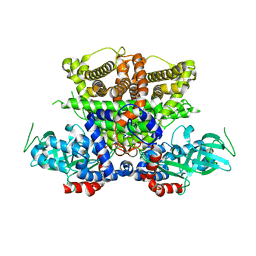 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | 分子名称: | Acyl-coenzyme A oxidase | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3I
 
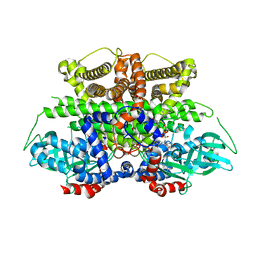 | | Crystal structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans complexed with FAD and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.683 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3G
 
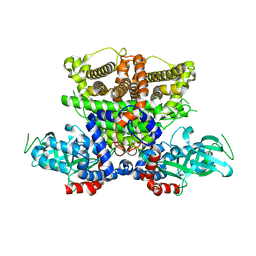 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | 分子名称: | Acyl-coenzyme A oxidase | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HI5
 
 | |
8HI6
 
 | |
5K3J
 
 | | Crystals structure of Acyl-CoA oxidase-2 in Caenorhabditis elegans bound with FAD, ascaroside-CoA, and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2GS7
 
 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | 分子名称: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | 著者 | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | 登録日 | 2006-04-25 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
4O9F
 
 | |
2GS2
 
 | | Crystal Structure of the active EGFR kinase domain | | 分子名称: | Epidermal growth factor receptor | | 著者 | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | 登録日 | 2006-04-25 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor.
Cell(Cambridge,Mass.), 125, 2006
|
|
4O9L
 
 | |
4JDL
 
 | |
4JDA
 
 | | Complex structure of abscisic acid receptor PYL3 with (-)-ABA | | 分子名称: | (2Z,4E)-5-[(1R)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | 著者 | Zhang, X, Wang, G, Chen, Z. | | 登録日 | 2013-02-24 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
4OA7
 
 | | Crystal structure of Tankyrase1 in complex with IWR1 | | 分子名称: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | 著者 | Zhang, X, He, H. | | 登録日 | 2014-01-03 | | 公開日 | 2015-01-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
3IYL
 
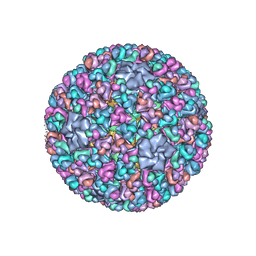 | | Atomic CryoEM Structure of a Nonenveloped Virus Suggests How Membrane Penetration Protein is Primed for Cell Entry | | 分子名称: | Core protein VP6, MYRISTIC ACID, Outer capsid VP4, ... | | 著者 | Zhang, X, Jin, L, Fang, Q, Hui, W, Zhou, Z.H. | | 登録日 | 2010-02-02 | | 公開日 | 2010-05-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry.
Cell(Cambridge,Mass.), 141, 2010
|
|
2MX6
 
 | |
5T2C
 
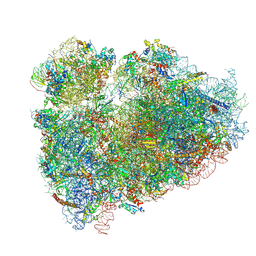 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Zhang, X, Lai, M, Zhou, Z.H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
