3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
5IC9
 
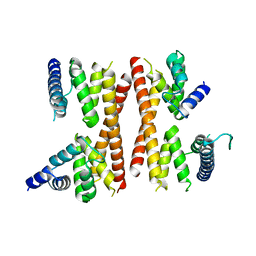 | | Structure of the CTD complex of Utp12 and Utp13 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC8
 
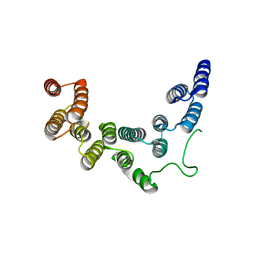 | | Structure of UTP6 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
7RIK
 
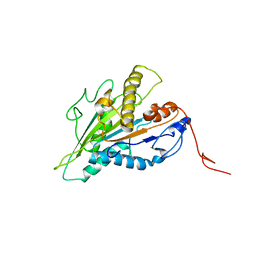 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | Descriptor: | Kinesin-1 heavy chain | | Authors: | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
7V4Q
 
 | |
5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | Descriptor: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
2I6K
 
 | | Crystal structure of human type I IPP isomerase complexed with a substrate analog | | Descriptor: | ACETIC ACID, AMINOETHANOLPYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase 1, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
5ICA
 
 | | Structure of the CTD complex of UTP12, Utp13, Utp1 and Utp21 | | Descriptor: | Periodic tryptophan protein 2-like protein, Putative U3 snoRNP protein, Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.507 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
8EPB
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*A)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*T)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPF
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*TP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
6KIK
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | Descriptor: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIY
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KY6
 
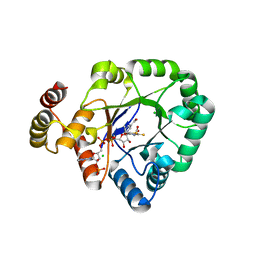 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | Descriptor: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6E9F
 
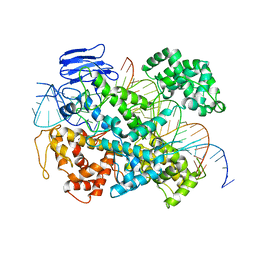 | | EsCas13d-crRNA-target RNA ternary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, RNA (27-MER), ... | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
8EPI
 
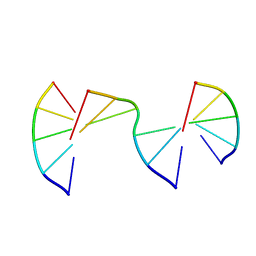 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*TP*A)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
7V4R
 
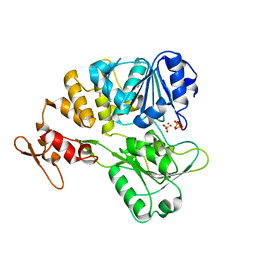 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
4NSX
 
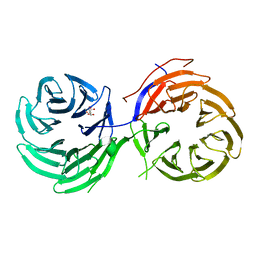 | | Crystal Structure of the Utp21 tandem WD Domain | | Descriptor: | CITRATE ANION, U3 small nucleolar RNA-associated protein 21 | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Utp21 Tandem WD Domain Provides Insight into the Organization of the UTPB Complex Involved in Ribosome Synthesis
Plos One, 9, 2014
|
|
8WCS
 
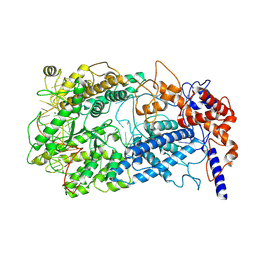 | | Cryo-EM structure of Cas13h1-crRNA binary complex | | Descriptor: | 66-nt crRNA, Cas13h1, MAGNESIUM ION | | Authors: | Zhang, C. | | Deposit date: | 2023-09-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism for target RNA recognition and cleavage of Cas13h.
Nucleic Acids Res., 52, 2024
|
|
8YA4
 
 | |
8Y9W
 
 | |
8J8D
 
 | |
8J8T
 
 | |
5GP1
 
 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
8EPG
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPE
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
