6LW0
 
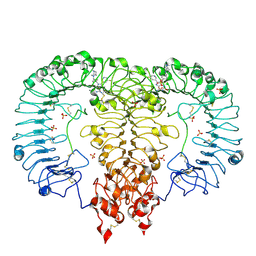 | | Crystal structure of TLR7/Cpd-6 (DSR-139293) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purin-6-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6ECS
 
 | |
6LVZ
 
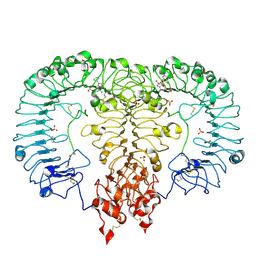 | | Crystal structure of TLR7/Cpd-3 (SM-394830) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(pyridin-3-ylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6EDJ
 
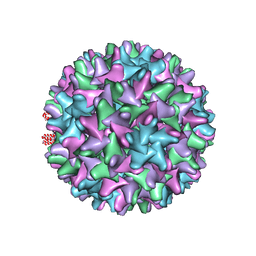 | | Cryo-EM structure of Woodchuck hepatitis virus capsid | | Descriptor: | External core antigen | | Authors: | Zhao, Z, Wang, J.C, Zlotnick, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Structural Differences between the Woodchuck Hepatitis Virus Core Protein in the Dimer and Capsid States Are Consistent with Entropic and Conformational Regulation of Assembly.
J.Virol., 93, 2019
|
|
8GXN
 
 | | The crystal structure of CsFAOMT2 in complex with SAH | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, caffeyl-CoA-O-methyltransferase | | Authors: | Zhang, Z.M, Zhou, Y.E, Huang, H.S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Characterization of two O-methyltransferases involved in the biosynthesis of O-methylated catechins in tea plant.
Nat Commun, 14, 2023
|
|
8GXO
 
 | |
8K53
 
 | |
8F3E
 
 | | Trimer of aminoglycoside efflux pump AcrD | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity.
Mbio, 14, 2023
|
|
8EOJ
 
 | | Microsomal triglyceride transfer protein | | Descriptor: | Microsomal triglyceride transfer protein large subunit, Protein disulfide-isomerase | | Authors: | Zhang, Z. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8ENE
 
 | |
8EOR
 
 | | Liver carboxylesterase 1 | | Descriptor: | ETHYL ACETATE, Liver carboxylesterase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, Z, Yu, E. | | Deposit date: | 2022-10-04 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
4YJL
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A2 | | Descriptor: | 1,2-ETHANEDIOL, APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
1BCC
 
 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
7Y0C
 
 | | Crystal structure of BD55-1403 and SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-1403 Fab heavy chain, BD55-1403 Fab light chain, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
8JF3
 
 | | C-Src in complex with compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Zhang, Z.M, Huang, H.S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.84647632 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3D8X
 
 | | Crystal Structure of Saccharomyces cerevisiae NDPPH Dependent Thioredoxin Reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Zhang, Z.Y, Bao, R, Yu, J, Chen, Y.X, Zhou, C.-Z. | | Deposit date: | 2008-05-26 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin
Biochim.Biophys.Acta, 1794, 2009
|
|
6LVX
 
 | | Crystal structure of TLR7/Cpd-1 (SM-374527) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-butoxy-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
4KZG
 
 | |
4YJE
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A1 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
5KLQ
 
 | | Crystal structure of HopZ1a in complex with IP6 and CoA | | Descriptor: | CITRIC ACID, COENZYME A, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
5KLP
 
 | | Crystal structure of HopZ1a in complex with IP6 | | Descriptor: | CITRIC ACID, INOSITOL HEXAKISPHOSPHATE, Orf34 | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-08-10 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4YK6
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A4 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
6K0Z
 
 | | Substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, GLYCEROL | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4967258 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
3VAX
 
 | | Crystal structure of DndA from streptomyces lividans | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein dndA | | Authors: | Zhang, Z, Chen, F, Lin, K, Wu, G. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation
Plos One, 7, 2012
|
|
