4XPU
 
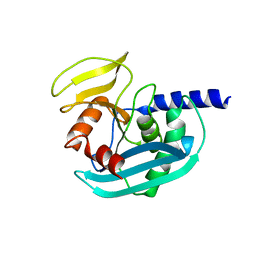 | | The crystal structure of EndoV from E.coli | | Descriptor: | Endonuclease V | | Authors: | Xie, W, Zhang, Z. | | Deposit date: | 2015-01-18 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of E. coli endonuclease V, an essential enzyme for deamination repair
Sci Rep, 5, 2015
|
|
4XBK
 
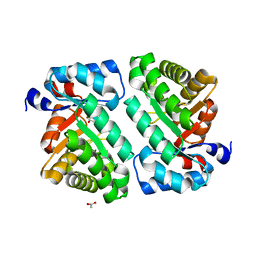 | | 2-deoxyribose-5-phosphate aldolase from Lactobacillus brevis | | Descriptor: | ACETIC ACID, Deoxyribose-phosphate aldolase | | Authors: | Jiao, X.-C, Pan, J, Xu, G.-C, Kong, X.-D, Chen, Q, Zhang, Z.-J, Xu, J.-H. | | Deposit date: | 2014-12-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis
Catalysis Science And Technology, 5, 2015
|
|
7YE6
 
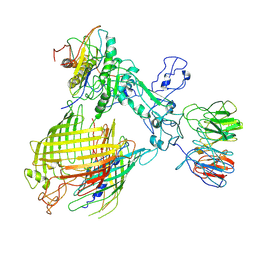 | | BAM-EspP complex structure with BamA-N427C/EspP-R1297C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
7YE4
 
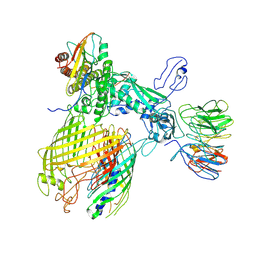 | | BAM-EspP complex structure with BamA-G431C and G781C/EspP-N1293C and A1043C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6BF8
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-03-30 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6B7Y
 
 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BFC
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF6
 
 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
8H7B
 
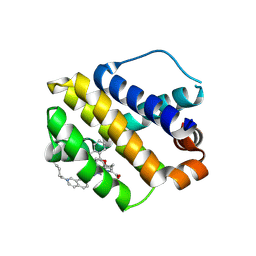 | | The crystal structure of human mcl1 kinase domain in complex with MCL1-M-EBA | | Descriptor: | 7-[3-(isoquinolin-7-yloxymethyl)-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46408451 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
6B3Q
 
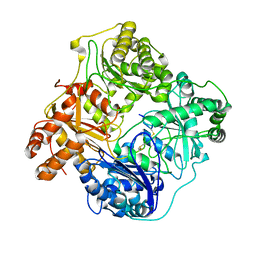 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
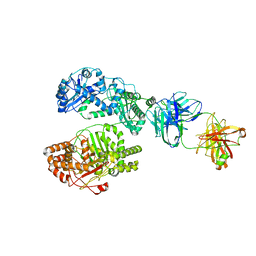 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | Descriptor: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-10 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5BX1
 
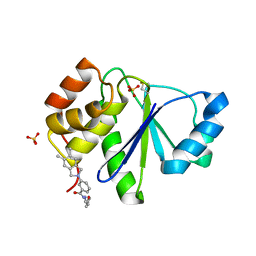 | | Crystal Structure of PRL-1 complex with compound analogy 3 | | Descriptor: | 3-(5,6-dimethyl-2H-isoindol-2-yl)-N'-[(E)-furan-2-ylmethylidene]benzohydrazide, Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Liu, S, Bai, Y, Zhang, Z. | | Deposit date: | 2015-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of PRL-1 complex with compound analogy 3
To Be Published
|
|
6JO8
 
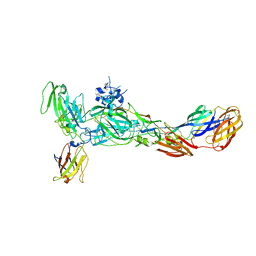 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
8PKP
 
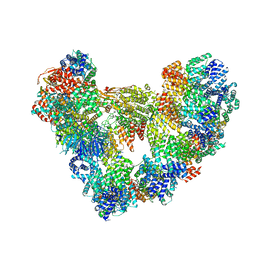 | | Cryo-EM structure of the apo Anaphase-promoting complex/cyclosome (APC/C) at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
8HOY
 
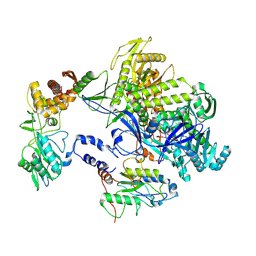 | | Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of DNA replication machinery from human monkeypox virus
To Be Published
|
|
6JX1
 
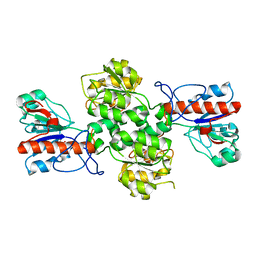 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JWG
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
3PS5
 
 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
3BLU
 
 | | crystal structure YopH complexed with inhibitor PVS | | Descriptor: | (ethenylsulfonyl)benzene, Tyrosine-protein phosphatase yopH | | Authors: | Liu, S, Zhang, Z.-Y. | | Deposit date: | 2007-12-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl vinyl sulfonates and sulfones as active site-directed and mechanism-based probes for protein tyrosine phosphatases
J.Am.Chem.Soc., 130, 2008
|
|
7D3Y
 
 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
