3LNL
 
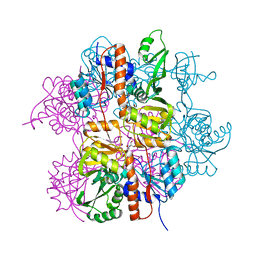 | | Crystal structure of Staphylococcus aureus protein SA1388 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UPF0135 protein SA1388, ZINC ION | | Authors: | Singh, K.S, Chruszcz, M, Zhang, X, Minor, W, Zhang, H. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Conserved Hypothetical Protein Sa1388 from S. aureus Reveals a Capped Hexameric Toroid with Two Pii Domain Lids and a Dinuclear Metal Center.
Bmc Struct.Biol., 6, 2006
|
|
8R64
 
 | |
5E6P
 
 | |
6MS7
 
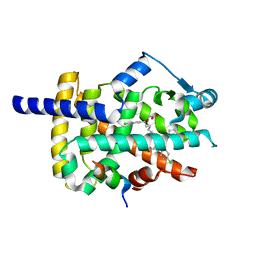 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8KGF
 
 | | Structure of AmCas12a with crRNA | | Descriptor: | CRISPR-associated endonuclease Cas12a, MAGNESIUM ION, RNA (44-MER) | | Authors: | Feng, Y, Zhang, X, Shi, J, Ma, P, Tang, J, Huang, X. | | Deposit date: | 2023-08-18 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A novel CRISPR/Cas12a in complex with a crRNA.
To Be Published
|
|
7SII
 
 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
8IAB
 
 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8QN4
 
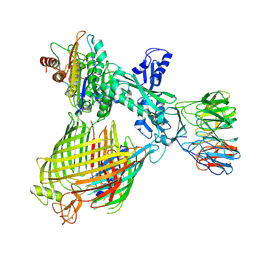 | | Structure of BAM-EspP complex in the non-closing EspP state | | Descriptor: | EspP epsilon, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Xie, T, Pang, J, Shen, C, Chang, S, Tang, X, Zhang, X, Dong, H, Zhou, R. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Dynamic topology-mediated maturation of beta-barrel proteins in BAM-catalyzed folding
To Be Published
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8RE4
 
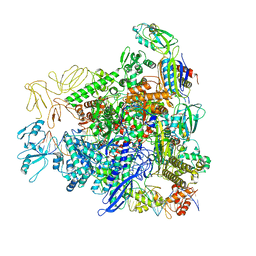 | |
8RED
 
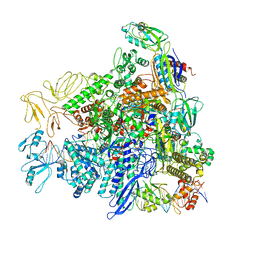 | |
8REE
 
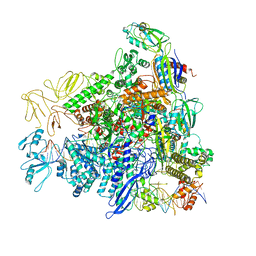 | |
8REB
 
 | |
8REC
 
 | |
8REA
 
 | |
1D59
 
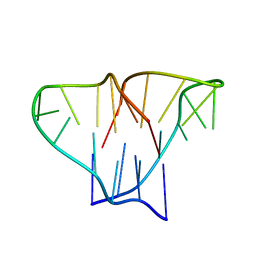 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1992-02-25 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
3LQ9
 
 | | Crystal structure of human REDD1, a hypoxia-induced regulator of mTOR | | Descriptor: | DNA-damage-inducible transcript 4 protein | | Authors: | Vega-Rubin-de-Celis, S, Abdallah, Z, Brugarolas, J, Zhang, X. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and functional implications of the negative mTORC1 regulator REDD1.
Biochemistry, 49, 2010
|
|
6NT9
 
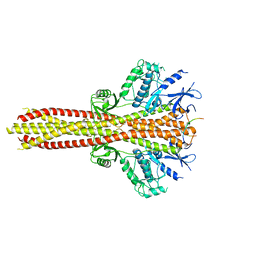 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | Descriptor: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
1DKZ
 
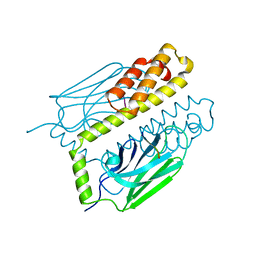 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 1 NATIVE CRYSTALS | | Descriptor: | SUBSTRATE BINDING DOMAIN OF DNAK, SUBSTRATE PEPTIDE (7 RESIDUES) | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
6LS3
 
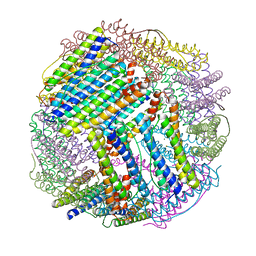 | |
6LS0
 
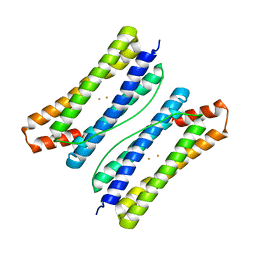 | |
8B4J
 
 | | Rfa1-N-terminal domain in complex with phosphorylated Ddc2 | | Descriptor: | 1,2-ETHANEDIOL, DNA damage checkpoint protein LCD1, Replication factor A protein 1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A DNA damage-induced phosphorylation circuit enhances Mec1 ATR Ddc2 ATRIP recruitment to Replication Protein A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4K
 
 | |
