6XOT
 
 | |
2AJ3
 
 | | Crystal Structure of a Cross-Reactive HIV-1 Neutralizing CD4-Binding Site Antibody Fab m18 | | Descriptor: | Fab m18, Heavy Chain, Light Chain, ... | | Authors: | Prabakaran, P, Gan, J, Wu, Y.Q, Zhang, M.Y, Dimitrov, D.S, Ji, X. | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural mimicry of CD4 by a cross-reactive HIV-1 neutralizing antibody with CDR-H2 and H3 containing unique motifs.
J.Mol.Biol., 357, 2006
|
|
5KJ7
 
 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from XFEL diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
7LN4
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMZ
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMY
 
 | | Cryo-EM structure of human p97 in complex with NMS-873 in the presence of ATP, Npl4/Ufd1, and Ub6 | | Descriptor: | 3-[3-cyclopentylsulfanyl-5-[[3-methyl-4-(4-methylsulfonylphenyl)phenoxy]methyl]-1,2,4-triazol-4-yl]pyridine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN2
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN0
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN6
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 2, Open State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN5
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 1, Close State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN1
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN3
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MEX
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
5KJ8
 
 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from synchrotron diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | Descriptor: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | Authors: | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | Descriptor: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5CCH
 
 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5CCJ
 
 | |
5CCG
 
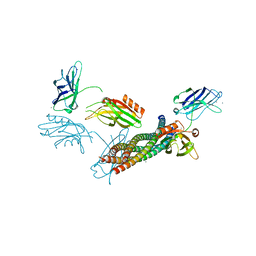 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
6VHH
 
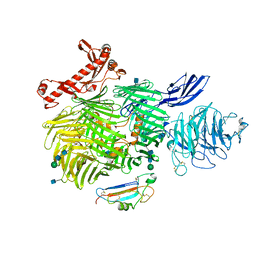 | | Human Teneurin-2 and human Latrophilin-3 binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L3, ... | | Authors: | Xie, Y, Li, J, Arac, D, Zhao, M. | | Deposit date: | 2020-01-09 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism.
Nat Commun, 11, 2020
|
|
8F4H
 
 | | RT XFEL structure of Photosystem II 1200 microseconds after the third illumination at 2.10 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4F
 
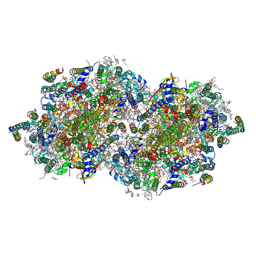 | | RT XFEL structure of Photosystem II 500 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4C
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8EZ5
 
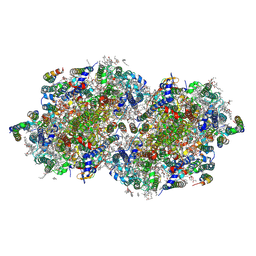 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4K
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.16 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
