7WWZ
 
 | | BRD4-BD1 complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
2D82
 
 | | Target Structure-Based Discovery of Small Molecules that Block Human p53 and CREB Binding Protein (CBP) Association | | Descriptor: | 9-ACETYL-2,3,4,9-TETRAHYDRO-1H-CARBAZOL-1-ONE, CREB-binding protein | | Authors: | Sachchidanand, Resnick-Silverman, L, Yan, S, Mujtaba, S, Liu, W.J, Zeng, L, Manfredi, J.J, Zhou, M.M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Target structure-based discovery of small molecules that block human p53 and CREB binding protein association
Chem.Biol., 13, 2006
|
|
1JM4
 
 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
1R4K
 
 | | Solution Structure of the Drosophila Argonaute 1 PAZ Domain | | Descriptor: | Argonaute 1 | | Authors: | Yan, K.S, Yan, S, Farooq, A, Han, A, Zeng, L, Zhou, M.-M. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and conserved RNA binding of the PAZ domain
Nature, 426, 2003
|
|
1XR0
 
 | | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors | | Descriptor: | Basic fibroblast growth factor receptor 1, FGFR signalling adaptor SNT-1 | | Authors: | Dhalluin, C, Yan, K.S, Plotnikova, O, Lee, K.W, Zeng, L, Kuti, M, Mujtaba, S, Goldfarb, M.P, Zhou, M.-M. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors
Mol.Cell, 6, 2000
|
|
1SZV
 
 | | Structure of the Adaptor Protein p14 reveals a Profilin-like Fold with Novel Function | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein | | Authors: | Qian, C, Zhang, Q, Wang, X, Zeng, L, Farooq, A, Zhou, M.M. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Adaptor Protein p14 Reveals a Profilin-like Fold with Distinct Function
J.Mol.Biol., 347, 2005
|
|
2AQF
 
 | | Structural and functional analysis of ADA2 alpha swirm domain | | Descriptor: | transcriptional adaptor 2, Ada2 alpha | | Authors: | Qian, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and chromosomal DNA binding of the SWIRM domain.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2AQE
 
 | | Structural and functional analysis of ada2 alpha swirm domain | | Descriptor: | Transcriptional adaptor 2, Ada2 alpha | | Authors: | Qian, C, Zhang, Q, Zeng, L, Zhou, M.-M. | | Deposit date: | 2005-08-17 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and chromosomal DNA binding of the SWIRM domain
Nat.Struct.Mol.Biol., 12, 2005
|
|
7XFG
 
 | |
7XEZ
 
 | |
7Y0I
 
 | | Solution structures of ASH1L PHD domain in complex with H3K4me2 peptide | | Descriptor: | ALA-ARG-THR-MLY-GLN-THR-ALA-ARG-LYS-SER-THR-GLY-GLY-LYS-ALA, Histone-lysine N-methyltransferase ASH1L, ZINC ION | | Authors: | Yu, M, Zeng, L. | | Deposit date: | 2022-06-05 | | Release date: | 2022-10-12 | | Method: | SOLUTION NMR | | Cite: | Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer.
Front Oncol, 12, 2022
|
|
3LJW
 
 | | Crystal Structure of the Second Bromodomain of Human Polybromo | | Descriptor: | ACETATE ION, Protein polybromo-1, SODIUM ION | | Authors: | Charlop-Powers, Z, Zhou, M.M, Zeng, L, Zhang, Q. | | Deposit date: | 2010-01-26 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
Cell Res., 20, 2010
|
|
8TUM
 
 | |
8TUX
 
 | |
8TUW
 
 | |
1JSP
 
 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
1N3J
 
 | | Structure and Substrate of a Histone H3 Lysine Methyltransferase from Paramecium Bursaria Chlorella Virus 1 | | Descriptor: | Histone H3 Lysine Methyltransferase | | Authors: | Manzur, K.L, Farooq, A, Zeng, L, Plotnikova, O, Sachchidanand, Koch, A.W, Zhou, M.-M. | | Deposit date: | 2002-10-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3.
Nat.Struct.Biol., 10, 2003
|
|
1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
1OP4
 
 | | Solution Structure of Neural Cadherin Prodomain | | Descriptor: | Neural-cadherin | | Authors: | Koch, A.W, Farooq, A, Shan, W, Zeng, L, Colman, D.R, Zhou, M.-M. | | Deposit date: | 2003-03-04 | | Release date: | 2004-03-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neural (N-) Cadherin Prodomain Reveals a Cadherin Extracellular Domain-like Fold without Adhesive Characteristics
Structure, 12, 2004
|
|
1OY2
 
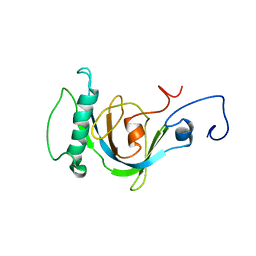 | | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | | Descriptor: | SHC transforming protein | | Authors: | Farooq, A, Zeng, L, Yan, K.S, Ravichandran, K.S, Zhou, M.-M. | | Deposit date: | 2003-04-03 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc
Structure, 11, 2003
|
|
4LGB
 
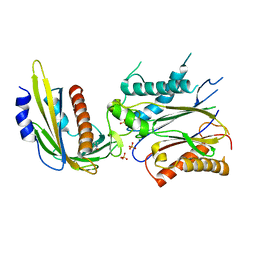 | | ABA-mimicking ligand N-(1-METHYL-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-(4-METHYLPHENYL)METHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LG5
 
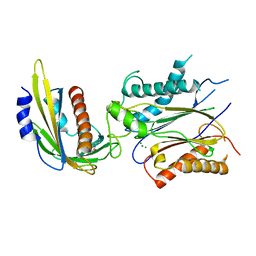 | | ABA-mimicking ligand QUINABACTIN in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LGA
 
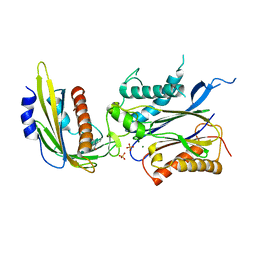 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
1N3H
 
 | | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | | Descriptor: | SHC Transforming protein | | Authors: | Farooq, A, Zeng, L, Yan, K.S, Ravichandran, K.S, Zhou, M.-M. | | Deposit date: | 2002-10-28 | | Release date: | 2003-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc
Structure, 11, 2003
|
|
2KFT
 
 | |
