6VX9
 
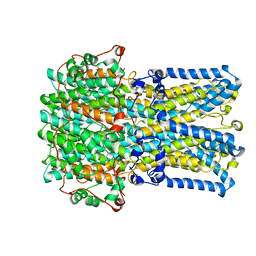 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
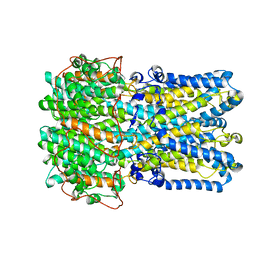 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
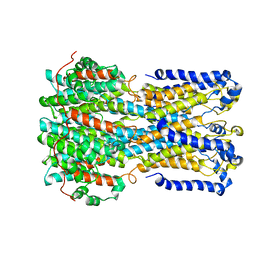 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6J20
 
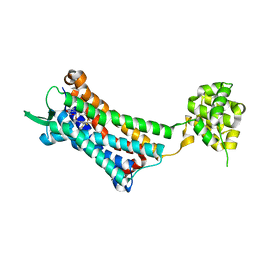 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6J21
 
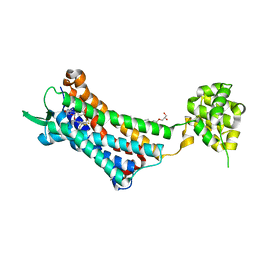 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
5TGZ
 
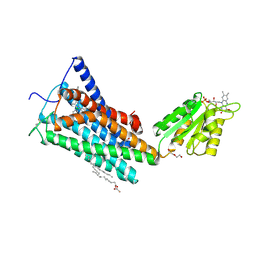 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
3E51
 
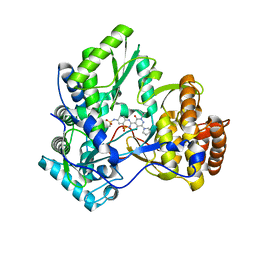 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-pyrrolidin-1-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 5: Exploration of pyridazinones containing 6-amino-substituents.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6IIV
 
 | | Crystal structure of the human thromboxane A2 receptor bound to daltroban | | Descriptor: | 2-[4-[2-[(4-chlorophenyl)sulfonylamino]ethyl]phenyl]ethanoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | Fan, H, Zhao, Q, Wu, B. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for ligand recognition of the human thromboxane A2receptor.
Nat. Chem. Biol., 15, 2019
|
|
1GA5
 
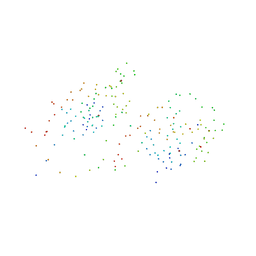 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*(5IT)P*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
8HZC
 
 | |
8HZB
 
 | |
8HZS
 
 | |
3CWJ
 
 | | Crystal structure of hcv ns5b polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-04-21 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-(1,1-Dioxo-1,4-dihydro-1lambda6-benzo[1,4]thiazin-3-yl)-5-hydroxy-2H-pyridazin-3-ones as potent inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CO9
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1HLZ
 
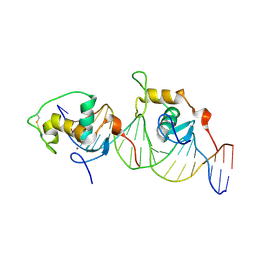 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
2ETT
 
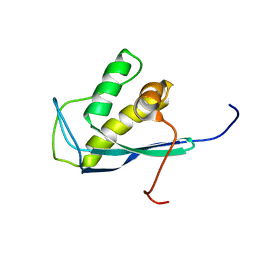 | | Solution Structure of Human Sorting Nexin 22 PX Domain | | Descriptor: | Sorting nexin-22 | | Authors: | Song, J, Zhao, Q, Tyler, R.C, Lee, M.S, Newman, C.L, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human sorting nexin 22.
Protein Sci., 16, 2007
|
|
8ILF
 
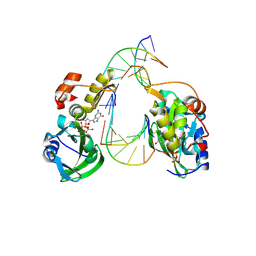 | | The crystal structure of dGTPalphaSe-Sp:DNApre-II:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), MANGANESE (II) ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Zhao, Q.W, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
3A2A
 
 | | The structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1 | | Descriptor: | CHLORIDE ION, Voltage-gated hydrogen channel 1 | | Authors: | Li, S.J, Unno, H, Zhou, Q, Zhao, Q, Zhai, Y, Sun, F. | | Deposit date: | 2009-05-08 | | Release date: | 2010-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1.
J.Biol.Chem., 285, 2010
|
|
3D28
 
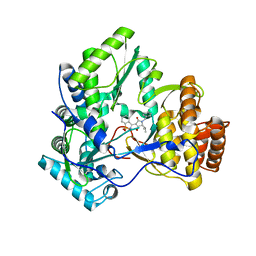 | | Crystal structure of hcv ns5b polymerase with a novel benzisothiazole inhibitor | | Descriptor: | (5S)-1-benzyl-3-(1,1-dioxido-1,2-benzisothiazol-3-yl)-4-hydroxy-5-(1-methylethyl)-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-05-07 | | Release date: | 2009-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of 1,1-dioxoisothiazole and benzo[b]thiophene-1,1-dioxide derivatives as novel inhibitors of hepatitis C virus NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BSA
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 2-({3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-(1,3-thiazol-5-yl)-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothia diazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1XSD
 
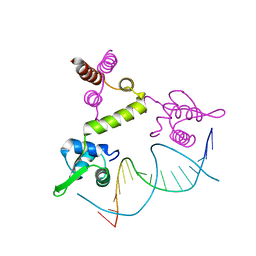 | | Crystal structure of the BlaI repressor in complex with DNA | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', penicillinase repressor | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Robinson, H, Scarsdale, N, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2004-10-19 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1TIZ
 
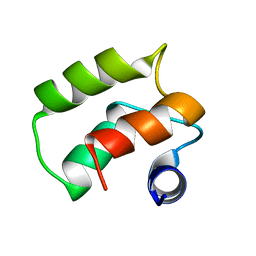 | | Solution Structure of a Calmodulin-Like Calcium-Binding Domain from Arabidopsis thaliana | | Descriptor: | calmodulin-related protein, putative | | Authors: | Song, J, Zhao, Q, Thao, S, Frederick, R.O, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: Solution structure of a calmodulin-like calcium-binding domain from Arabidopsis thaliana
J.Biomol.NMR, 30, 2004
|
|
1XRX
 
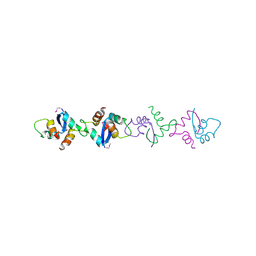 | | Crystal structure of a DNA-binding protein | | Descriptor: | CALCIUM ION, SeqA protein | | Authors: | Guarne, A, Brendler, T, Zhao, Q, Ghirlando, R, Austin, S, Yang, W. | | Deposit date: | 2004-10-16 | | Release date: | 2005-05-10 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization.
Embo J., 24, 2005
|
|
4TNX
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dGTP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TO3
 
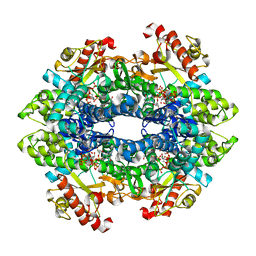 | | Structural basis of cellular dNTP regulation, SAMHD1-dGTP-dGTP-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
