7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ELK
 
 | |
9IKC
 
 | | Orf virus scaffolding protein Orfv075 | | Descriptor: | 62 kDa protein | | Authors: | Hyun, J, Kim, S, Ko, S, Kim, M, Jang, Y. | | Deposit date: | 2024-06-27 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Cryo-EM structure of orf virus scaffolding protein orfv075.
Biochem.Biophys.Res.Commun., 728, 2024
|
|
2C7B
 
 | | The Crystal Structure of EstE1, a New Thermophilic and Thermostable Carboxylesterase Cloned from a Metagenomic Library | | Descriptor: | CARBOXYLESTERASE | | Authors: | Byun, J.-S, Rhee, J.-K, Kim, D.-U, Oh, J.-W, Cho, H.-S. | | Deposit date: | 2005-11-21 | | Release date: | 2005-12-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Hyperthermophilic Esterase Este1 and the Relationship between its Dimerization and Thermostability Properties.
Bmc Struct.Biol., 7, 2007
|
|
2CKX
 
 | |
6IQ1
 
 | |
2M17
 
 | |
5BY2
 
 | |
5A2P
 
 | |
7WJ5
 
 | | Cryo-EM structure of human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand binding specificity | | Descriptor: | Gai1 antibody (scfv16), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Heo, Y.S, Yoon, E.J, Jeon, Y.E, Yun, J.-H, Ishimoto, N, Woo, H, Park, S.Y, Song, J, Lee, W.T. | | Deposit date: | 2022-01-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Elife, 11, 2022
|
|
6JCG
 
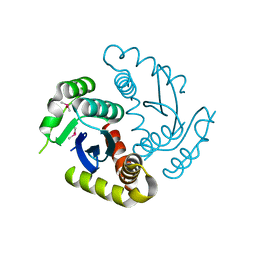 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
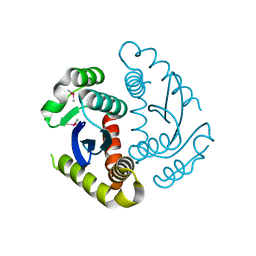 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6KHF
 
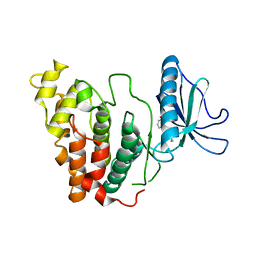 | | Crystal structure of CLK3 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KHE
 
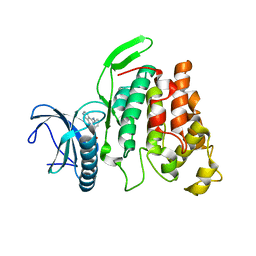 | | Crystal structure of CLK2 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KHD
 
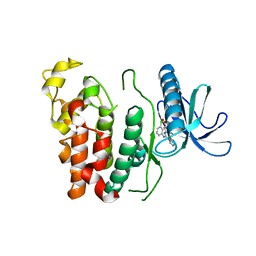 | | Crystal structure of CLK1 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK1 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
5G1D
 
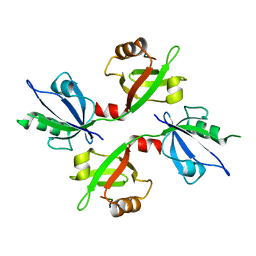 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNDECAN-4, SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5G1E
 
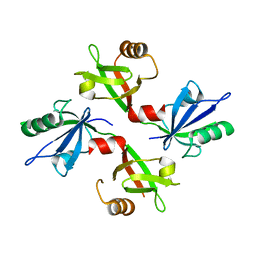 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
7FDJ
 
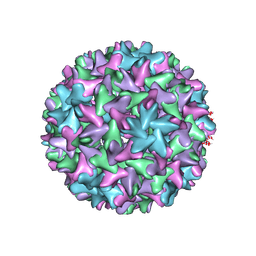 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EP6
 
 | | Engineered Hepatitis B virus core antigen T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EOY
 
 | | Engineered Hepatitis B virus core antigen T=3 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-24 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
6KVR
 
 | | Fatty acid amide hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Min, C.A, Yun, J.S, Chang, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Crystals, 9, 2019
|
|
8HU2
 
 | | Rattus Syntenin-1 PDZ domain with inhibitor | | Descriptor: | (2~{S})-2-(9~{H}-fluoren-9-ylmethoxycarbonylamino)-3-(4-oxidanylidene-5~{H}-pyrimidin-2-yl)propanoic acid, Syntenin-1 | | Authors: | Heo, Y, Lee, J, Yun, J.H, Lee, W. | | Deposit date: | 2022-12-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of STNPDZ with inhibitor at 1.60 Angstroms resolution.
To Be Published
|
|
