3B8K
 
 | | Structure of the Truncated Human Dihydrolipoyl Acetyltransferase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase | | Authors: | Yu, X, Hiromasa, Y, Tsen, H, Stoops, J.K, Roche, T.E, Zhou, Z.H. | | Deposit date: | 2007-11-01 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains
Structure, 16, 2008
|
|
9B2X
 
 | | Cryo-EM structure of Prefusion RSV F (RSV220975) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Yu, X, Langedijk, J.P.M. | | Deposit date: | 2024-03-17 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | An efficacious foldon-free prefusion F trimer vaccine for Respiratory Syncytial Virus by stabilizing trimerization-induced regions of instability
To Be Published
|
|
4ZS6
 
 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
2QCT
 
 | | Structure of Lyp with inhibitor I-C11 | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXY-3-{(4R)-1-[4-(1-NAPHTHYLAMINO)-4-OXOBUTYL]-1,2,3-TRIAZOLIDIN-4-YL}-1-BENZOFURAN-5-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Yu, X, Sun, J.P, Zhang, Z.Y. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Lyp and its complex with a selective inhibitor
To be Published
|
|
2REC
 
 | |
3OMH
 
 | | Crystal structure of PTPN22 in complex with SKAP-HOM pTyr75 peptide | | Descriptor: | Src kinase-associated phosphoprotein 2, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Yu, X, Sun, J.-P, Zhang, S, Zhang, Z.-Y. | | Deposit date: | 2010-08-26 | | Release date: | 2011-06-29 | | Last modified: | 2011-09-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate Specificity of Lymphoid-specific Tyrosine Phosphatase (Lyp) and Identification of Src Kinase-associated Protein of 55 kDa Homolog (SKAP-HOM) as a Lyp Substrate.
J.Biol.Chem., 286, 2011
|
|
8SWK
 
 | | Cryo-EM structure of NLRP3 closed hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SXN
 
 | | Structure of NLRP3 and NEK7 complex | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6MPB
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
3CNF
 
 | |
3FCG
 
 | | Crystal Structure Analysis of the Middle Domain of the Caf1A Usher | | Descriptor: | CHLORIDE ION, F1 capsule-anchoring protein | | Authors: | Yu, X, Visweswaran, G.R, Duck, Z, Marupakula, S, MacIntyre, S, Knight, S, Zavialov, A.V. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Caf1A usher possesses a Caf1 subunit-like domain that is crucial for Caf1 fibre secretion
Biochem.J., 418, 2009
|
|
4BM7
 
 | | Crystal Structure of IgG Fc F241A mutant with native glycosylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, IG GAMMA-1 CHAIN C REGION | | Authors: | Yu, X, Baruah, K, Harvey, D.J, Vasiljevic, S, Alonzi, D.S, Song, B, Higgins, M.K, Bowden, T.A, Crispin, M, Scanlan, C.N. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Engineering Hydrophobic Protein-Carbohydrate Interactions to Fine-Tune Monoclonal Antibodies.
J.Am.Chem.Soc., 135, 2013
|
|
8E76
 
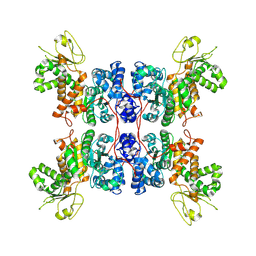 | | Cryo-EM structure of Apo form ME3 | | Descriptor: | NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8E78
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8E8O
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
3OLR
 
 | |
8FU7
 
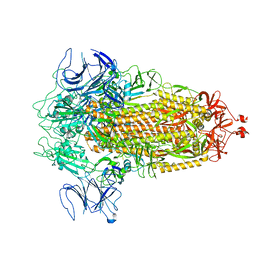 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
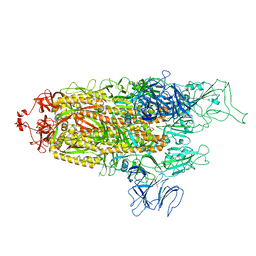 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
3IZX
 
 | | 3.1 Angstrom cryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Capsid protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Yu, X, Ge, P, Jiang, J, Atanasov, I, Zhou, Z.H. | | Deposit date: | 2011-01-15 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses.
Structure, 19, 2011
|
|
3J2V
 
 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
3J1R
 
 | | Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-terminal Type IV Pilin Helices | | Descriptor: | archaeal adhesion filament core | | Authors: | Yu, X, Goforth, C, Meyer, C, Rachel, R, Wirth, R, Schroeder, G.F, Egelman, E.H. | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-Terminal Type IV Pilin Helices.
J.Mol.Biol., 422, 2012
|
|
