5EM6
 
 | |
5HIE
 
 | | BRAF Kinase domain b3aC loop deletion mutant in complex with dabrafenib | | Descriptor: | Dabrafenib, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
7RR3
 
 | | Structure of Deep-Sea Phage NrS-1 Primase-Polymerase N300 in complex with calcium and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, CALCIUM ION, Primase | | Authors: | Wang, L, Yu, C, Sliz, P. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Molecular Dissection of the Primase and Polymerase Activities of Deep-Sea Phage NrS-1 Primase-Polymerase.
Front Microbiol, 12, 2021
|
|
5EM5
 
 | |
7RR4
 
 | |
5EDP
 
 | | EGFR kinase (T790M/L858R) apo | | Descriptor: | Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HIC
 
 | | EGFR kinase domain mutant "TMLR" with a imidazopyridinyl-aminopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-{2-[1-(cyclopropylsulfonyl)-1H-pyrazol-4-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
6XNN
 
 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
5EM8
 
 | |
5EDR
 
 | | EGFR kinase (T790M/L858R) with inhibitor compound 27: ~{N}-(1~{H}-indazol-3-yl)-7,7-dimethyl-2-(2-methylpyrazol-3-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Descriptor: | Epidermal growth factor receptor, ~{N}-(1~{H}-indazol-3-yl)-7,7-dimethyl-2-(2-methylpyrazol-3-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HCX
 
 | | EGFR kinase domain mutant "TMLR" with azabenzimidazole compound 7 | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]-2-methyl-1-propan-2-yl-imidazo[4,5-c]pyridin-6-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Noncovalent, Mutant-Selective Epidermal Growth Factor Receptor Inhibitor.
J.Med.Chem., 59, 2016
|
|
4D8O
 
 | |
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6U7V
 
 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
2MYP
 
 | |
2MYT
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
5XPY
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
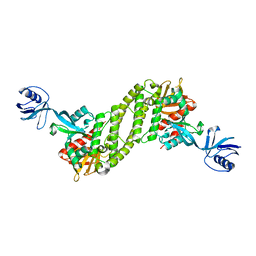 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7Y5V
 
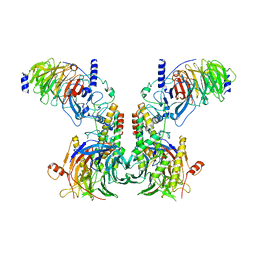 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y61
 
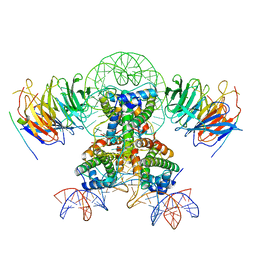 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y60
 
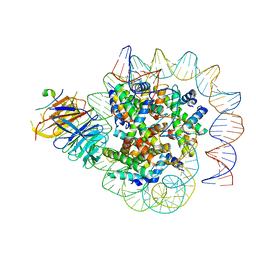 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5W
 
 | | Cryo-EM structure of the left-handed Di-tetrasome | | Descriptor: | Histone H3.1, Histone H4, Widom 601 DNA (147-MER) | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5U
 
 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
2MYN
 
 | | An arsenate reductase in reduced state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
8F41
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
