6J9F
 
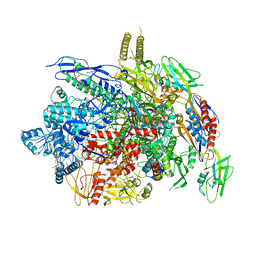 | |
6J9E
 
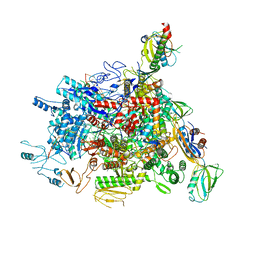 | |
8GZH
 
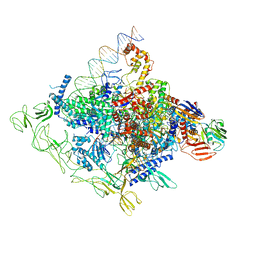 | |
6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | 分子名称: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFZ
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR2-ssDNA complex | | 分子名称: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFN
 
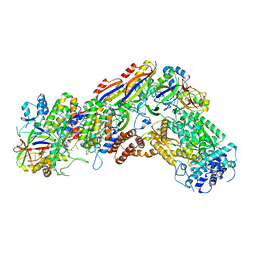 | | Crystal structure of Type III-A CRISPR Csm complex | | 分子名称: | MANGANESE (II) ION, RNA (32-MER), Type III-A CRISPR-associated RAMP protein Csm3, ... | | 著者 | You, L, Wang, J, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFU
 
 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | 分子名称: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | 分子名称: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | 分子名称: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
7YPB
 
 | |
7YPA
 
 | |
7YP9
 
 | |
6C5N
 
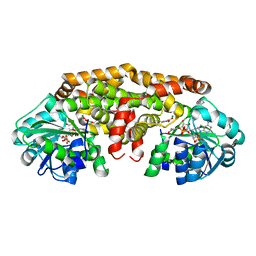 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | 分子名称: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | 著者 | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | 登録日 | 2018-01-16 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.673 Å) | | 主引用文献 | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | 分子名称: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | 分子名称: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GZG
 
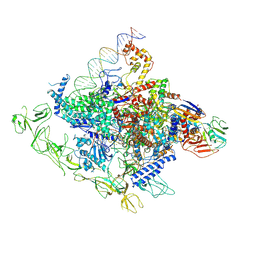 | |
7F0R
 
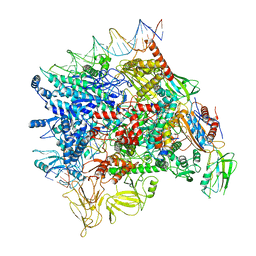 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | 分子名称: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2021-06-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7RDU
 
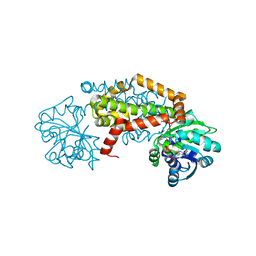 | |
6BUL
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 2 | | 分子名称: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McFeary, R.P. | | 登録日 | 2017-12-10 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6C55
 
 | | Crystal structure of Staphylococcus aureus Ketol-acid Reductosimerrase with hydroxyoxamate inhibitor 3 | | 分子名称: | (cyclohexylamino)(oxo)acetic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | 著者 | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schmbri, M, McGeary, R.P. | | 登録日 | 2018-01-14 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | 分子名称: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | 著者 | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | 登録日 | 2017-06-30 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2021-09-10 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
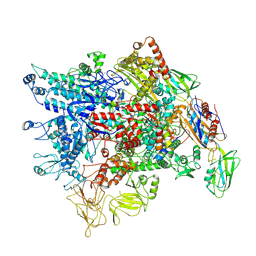 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2022-04-21 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-03 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
