4JQA
 
 | | AKR1C2 complex with mefenamic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, Aldo-keto reductase family 1 member C2, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
4JTQ
 
 | |
4JQ1
 
 | | AKR1C2 complex with naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
6ZSL
 
 | | Crystal structure of the SARS-CoV-2 helicase at 1.94 Angstrom resolution | | Descriptor: | PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Arrowsmith, C.H, von Delft, F, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
6QNV
 
 | | Fibrinogen-like globe domain of Human Tenascin-C | | Descriptor: | Tenascin | | Authors: | Coker, J.A, Bezerra, G.A, Bradshaw, W.J, Zhang, M, Yosaatmadja, Y, Fernandez-Cid, A, Shrestha, L, Burgess-Brown, N, Gileadi, O, Arrowsmith, C.H, Bountra, C, Midwood, K.S, Yue, W.W, Marsden, B.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fibrinogen-like globe domain of Human Tenascin-C
To Be Published
|
|
4ZAU
 
 | | AZD9291 complex with wild type EGFR | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Squire, C.J, Yosaatmadja, Y, Flanagan, J.U, McKeage, M. | | Deposit date: | 2015-04-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed.
J.Struct.Biol., 192, 2015
|
|
4DZ5
 
 | |
7QGI
 
 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QIF
 
 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7NIO
 
 | | Crystal structure of the SARS-CoV-2 helicase APO form | | Descriptor: | SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7NNG
 
 | | Crystal structure of the SARS-CoV-2 helicase in complex with Z2327226104 | | Descriptor: | 1-(2-methylphenyl)-1,2,3-triazole-4-carboxylic acid, PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5E5Q
 
 | | Racemic snakin-1 in P21/c | | Descriptor: | Snakin-1 | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5Y
 
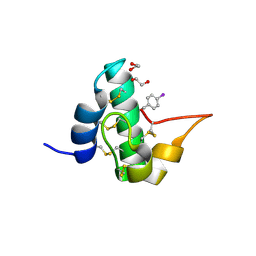 | | Quasi-racemic snakin-1 in P1 before radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5T
 
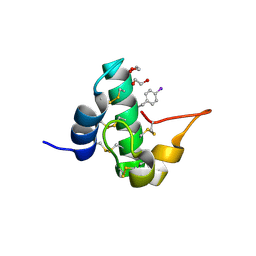 | | Quasi-racemic snakin-1 in P1 after radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7NN0
 
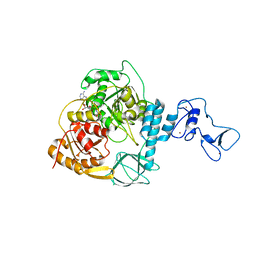 | | Crystal structure of the SARS-CoV-2 helicase in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4WRG
 
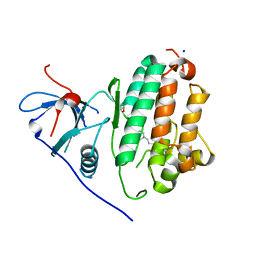 | |
4WRH
 
 | | AKR1C3 complexed with breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(aminomethyl)-2-chloro-6-methoxyphenoxy]-N-tert-butylacetamide, 5-methyl-4H-1,2,4-triazole-3-thiol, ... | | Authors: | Squire, C.J, Flanagan, J.U, Yosaatmadja, Y. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide trapped in active site of AKR1C3
To Be Published
|
|
4XBJ
 
 | | Y274F alanine racemase from E. coli inhibited by l-ala-p | | Descriptor: | Alanine racemase, biosynthetic, SULFATE ION, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
6C1O
 
 | | FGFR1 kinase domain complexed with FIIN-1 | | Descriptor: | Fibroblast growth factor receptor 1, N-(3-{[3-(2,6-dichloro-3,5-dimethoxyphenyl)-7-{[4-(diethylamino)butyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)prop-2-enamide, SULFATE ION | | Authors: | Kalyukina, M, Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A new class of FGFR1 inhibitors
To Be Published
|
|
5TCV
 
 | |
5TCW
 
 | |
3UWE
 
 | | AKR1C3 complexed with 3-phenoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-phenoxybenzoic acid, Aldo-keto reductase family 1 member C3, ... | | Authors: | Jackson, V.J, Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-12-01 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of AKR1C3 with 3-phenoxybenzoic acid bound
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6NVJ
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVG
 
 | | FGFR4 complex with N-(3,5-dichloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)phenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3,5-dichloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)phenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
