2RQR
 
 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | Descriptor: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | Authors: | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2DVW
 
 | |
2DZN
 
 | |
1HZD
 
 | | CRYSTAL STRUCTURE OF HUMAN AUH PROTEIN, AN RNA-BINDING HOMOLOGUE OF ENOYL-COA HYDRATASE | | Descriptor: | AU-BINDING PROTEIN/ENOYL-COA HYDRATASE | | Authors: | Kurimoto, K, Fukai, S, Nureki, O, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-01-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase.
Structure, 9, 2001
|
|
2JNS
 
 | |
3GFJ
 
 | |
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | Descriptor: | SOS 1 | | Authors: | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-02-18 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
2KHE
 
 | | Solution Structure of the Bacterial Toxin Rele from Thermus Thermophilus HB8 | | Descriptor: | Toxin-like protein | | Authors: | Suzuki, S, Kawazoe, M, Kaminishi, T, Takemoto, C, Muto, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacterial Toxin Rele from Thermus Thermophilus HB8
To be Published
|
|
1QQT
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
2EG9
 
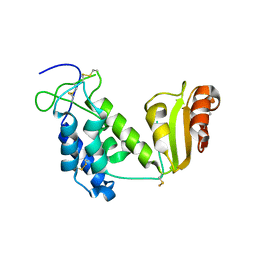 | | Crystal structure of the truncated extracellular domain of mouse CD38 | | Descriptor: | ADP-ribosyl cyclase 1 | | Authors: | Kukimoto-Niino, M, Mishima, C, Wakiyama, M, Terada, T, Shirouzu, M, Hara-Yokoyama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the truncated extracellular domain of mouse CD38
To be Published
|
|
4ENY
 
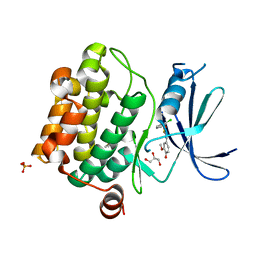 | | Crystal Structure of Pim-1 kinase in complex with (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-methoxybenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-methoxybenzylidene)-1,3-thiazolidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1Z54
 
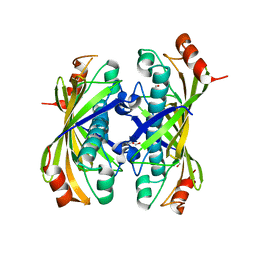 | | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus | | Descriptor: | GLYCEROL, probable thioesterase | | Authors: | Ihsanawati, Kaminishi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus
TO BE PUBLISHED
|
|
2EF1
 
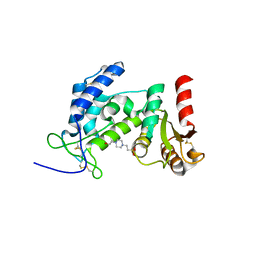 | | Crystal structure of the extracellular domain of human CD38 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADP-ribosyl cyclase 1 | | Authors: | Kukimoto-Niino, M, Nureki, O, Murayama, K, Shirouzu, M, Katada, T, Hara-Yokoyama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the extracellular domain of human CD38
To be Published
|
|
1ZPW
 
 | |
1ZRJ
 
 | | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c | | Descriptor: | E1B-55kDa-associated protein 5 isoform c | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c
To be Published
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
3KBB
 
 | | Crystal structure of putative beta-phosphoglucomutase from Thermotoga maritima | | Descriptor: | GLYCEROL, Phosphorylated carbohydrates phosphatase TM_1254, SULFATE ION | | Authors: | Strange, R.W, Antonyuk, S.V, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of a putative beta-phosphoglucomutase (TM1254) from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3JQM
 
 | | Binding of 5'-GTP to molybdenum cofactor biosynthesis protein MoaC from Thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5XVR
 
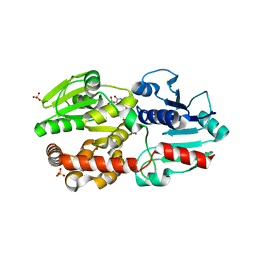 | |
3JQJ
 
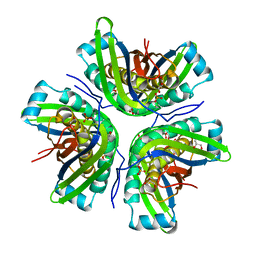 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | GLYCEROL, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KB6
 
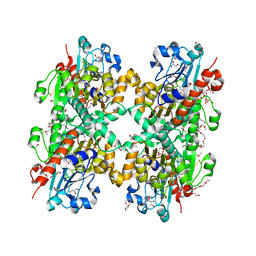 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4B04
 
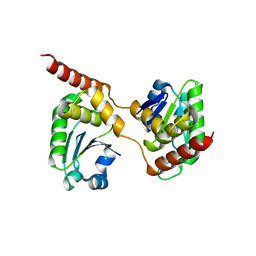 | | Crystal structure of the Catalytic Domain of Human DUSP26 (C152S) | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 26 | | Authors: | Won, E.-Y, Lee, D.Y, Park, S.G, Yokoyama, S, Kim, S.J, Chi, S.-W. | | Deposit date: | 2012-06-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | High-Resolution Crystal Structure of the Catalytic Domain of Human Dual-Specificity Phosphatase 26
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SUU
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-PUGNAc | | Descriptor: | Beta-hexosaminidase, SULFATE ION, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
5XHG
 
 | | Crystal structure of Trastuzumab Fab fragment bearing Ne-(o-azidobenzyloxycarbonyl)-L-lysine | | Descriptor: | (2-azidophenyl)methyl hydrogen carbonate, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
3SUW
 
 | |
