1L5Z
 
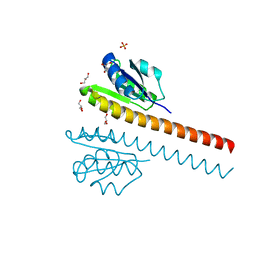 | | CRYSTAL STRUCTURE OF THE E121K SUBSTITUTION OF THE RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN DCTD, GLYCEROL, SULFATE ION | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
4ZK6
 
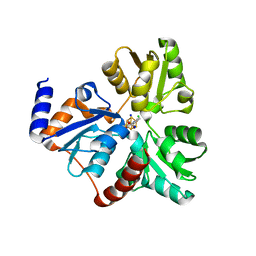 | | Crystallographic Capture of Quinolinate Synthase (NadA) from Pyrococcus horikoshii in its Substrates and Product-Bound States | | Descriptor: | ACETATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Saunders, A.H, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2015-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of Quinolinate Synthase from Pyrococcus horikoshii in the Presence of Its Product, Quinolinic Acid.
J.Am.Chem.Soc., 138, 2016
|
|
1GHB
 
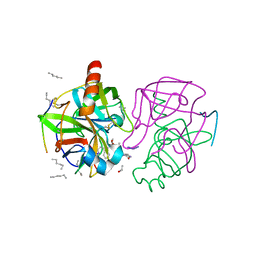 | | A SECOND ACTIVE SITE IN CHYMOTRYPSIN? THE X-RAY CRYSTAL STRUCTURE OF N-ACETYL-D-TRYPTOPHAN BOUND TO GAMMA-CHYMOTRYPSIN | | Descriptor: | ACETYL GROUP, GAMMA-CHYMOTRYPSIN, HEXANE, ... | | Authors: | Yennawar, H.P, Yennawar, N.H, Farber, G.K. | | Deposit date: | 1994-04-06 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A STRUCTURAL EXPLANATION FOR ENZYME MEMORY IN NONAQUEOUS SOLVENTS.
J.Am.Chem.Soc., 117, 1995
|
|
1GHA
 
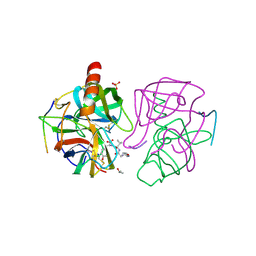 | | A SECOND ACTIVE SITE IN CHYMOTRYPSIN? THE X-RAY CRYSTAL STRUCTURE OF N-ACETYL-D-TRYPTOPHAN BOUND TO GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN A, ISOPROPYL ALCOHOL, PRO GLY VAL TYR PEPTIDE, ... | | Authors: | Yennawar, H.P, Yennawar, N.H, Farber, G.K. | | Deposit date: | 1994-04-06 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A STRUCTURAL EXPLANATION FOR ENZYME MEMORY IN NONAQUEOUS SOLVENTS.
J.Am.Chem.Soc., 117, 1995
|
|
6NSO
 
 | | An Unexpected Intermediate in the Reaction Catalyzed by Quinolinate Synthase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, IRON/SULFUR CLUSTER, Quinolinate synthase A | | Authors: | Esakova, O.A, Grove, T.L, Silakov, A, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
6NSU
 
 | | Crystallographic Capture of Quinolinate Synthase (NadA) from Pyrococcus horikoshii in its Substrates and Product-Bound States | | Descriptor: | DIDEHYDROASPARTATE, IRON/SULFUR CLUSTER, Quinolinate synthase A | | Authors: | Esakova, O.A, Grove, T.L, Silakov, A, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
6OR8
 
 | | An Unexpected Intermediate in the Reaction Catalyzed by Quinolinate Synthase | | Descriptor: | 2-hydroxy-N-[(1S)-1-hydroxy-3-oxopropyl]-L-aspartic acid, IRON/SULFUR CLUSTER, PHOSPHATE ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
4FER
 
 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose | | Descriptor: | ACETIC ACID, Expansin-yoaJ, GLYCEROL, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-05-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FFT
 
 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with mixed-linkage glucan | | Descriptor: | ACETIC ACID, Expansin-yoaJ, SULFATE ION, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-06-01 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7MJV
 
 | | MiaB in the complex with s-adenosylmethionine and RNA | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJY
 
 | | MiaB in the complex with s-adenosyl-L-homocysteine and RNA | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJZ
 
 | | The structure of MiaB with pentasulfide bridge | | Descriptor: | IRON/SULFUR CLUSTER, PENTASULFIDE-SULFUR, SODIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJW
 
 | | Methylated MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 5'-DEOXYADENOSINE, FE3-S4 methylated cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJX
 
 | | MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXYADENOSINE, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
4FG4
 
 | |
4FG2
 
 | |
