5XN2
 
 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
6KDN
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDM
 
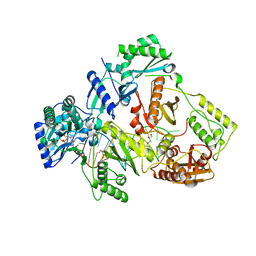 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:entecavir 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDJ
 
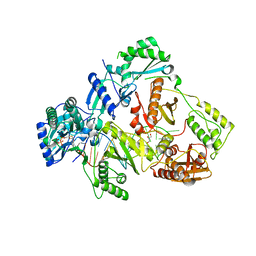 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KGA
 
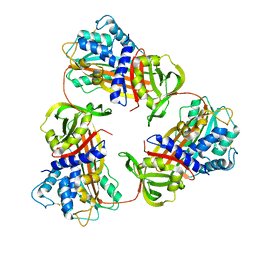 | |
6KDO
 
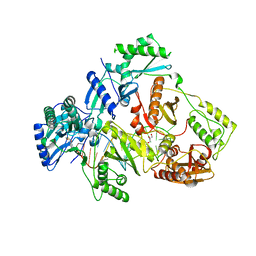 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
7COF
 
 | |
7COG
 
 | |
3WQC
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 | | Descriptor: | CHLORIDE ION, D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQD
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-erythro-3-hydroxyaspartate | | Descriptor: | (3S)-3-hydroxy-D-aspartic acid, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQF
 
 | |
3WQE
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-allothreonine | | Descriptor: | D-allothreonine, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQG
 
 | | D-threo-3-hydroxyaspartate dehydratase C353A mutant in the metal-free form | | Descriptor: | D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3VTZ
 
 | |
3VUN
 
 | | Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yasutake, Y, Suzuki, T, Kawaguchi, A, Nobusawa, E. | | Deposit date: | 2012-07-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group
To be Published
|
|
3WEC
 
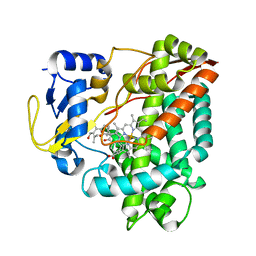 | | Structure of P450 RauA (CYP1050A1) complexed with a biosynthetic intermediate of aurachin RE | | Descriptor: | 3-[(2E,6E,9R)-9-hydroxy-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]-2-methylquinolin-4(1H)-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yasutake, Y, Kitagawa, W, Tamura, T. | | Deposit date: | 2013-07-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the quinoline N-hydroxylating cytochrome P450 RauA, an essential enzyme that confers antibiotic activity on aurachin alkaloids
Febs Lett., 588, 2014
|
|
2Z36
 
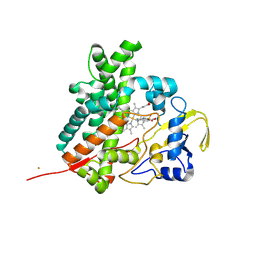 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
3A51
 
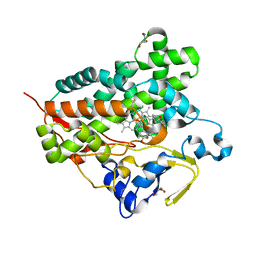 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound 25-hydroxyvitamin D3 | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4H
 
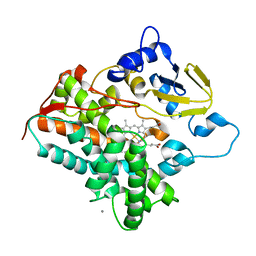 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (orthorhombic crystal form) | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4G
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (trigonal crystal form) | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4Z
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A50
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3B1Q
 
 | | Structure of Burkholderia thailandensis nucleoside kinase (BthNK) in complex with inosine | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETATE ION, INOSINE, ... | | Authors: | Yasutake, Y, Ota, H, Hino, E, Sakasegawa, S, Tamura, T. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Burkholderia thailandensis nucleoside kinase: implications for the catalytic mechanism and nucleoside selectivity
Acta Crystallogr.,Sect.D, 67, 2011
|
|
