5Z8L
 
 | |
5ZBH
 
 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
5ZBQ
 
 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
7VNB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7EAR
 
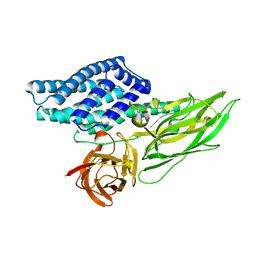 | | A positively charged mutant Cry3Aa endotoxin | | Descriptor: | Crystaline entomocidal protoxin | | Authors: | Yang, Z, Lee, M.M, Chan, M.K. | | Deposit date: | 2021-03-08 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficient intracellular delivery of p53 protein by engineered protein crystals restores tumor suppressing function in vivo.
Biomaterials, 271, 2021
|
|
8IAB
 
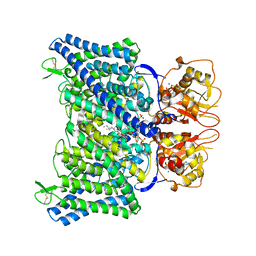 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
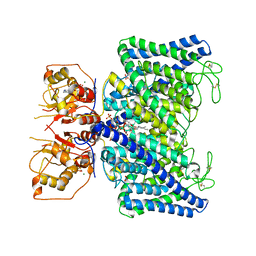 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
1EFL
 
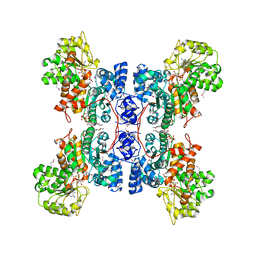 | | HUMAN MALIC ENZYME IN A QUATERNARY COMPLEX WITH NAD, MG, AND TARTRONATE | | Descriptor: | MAGNESIUM ION, MALIC ENZYME, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
1EI3
 
 | | CRYSTAL STRUCTURE OF NATIVE CHICKEN FIBRINOGEN | | Descriptor: | FIBRINOGEN | | Authors: | Yang, Z, Mochalkin, I, Veerapandian, L, Riley, M, Doolittle, R.F. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal structure of native chicken fibrinogen at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1GZ3
 
 | | Molecular mechanism for the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-14 | | Release date: | 2003-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
1EFK
 
 | | STRUCTURE OF HUMAN MALIC ENZYME IN COMPLEX WITH KETOMALONATE | | Descriptor: | ALPHA-KETOMALONIC ACID, MAGNESIUM ION, MALIC ENZYME, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
1H2H
 
 | | Crystal structure of TM1643 | | Descriptor: | HYPOTHETICAL PROTEIN TM1643, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Z, Savchenko, A, Edwards, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-15 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643.
J. Biol. Chem., 278, 2003
|
|
1GQ2
 
 | | Malic Enzyme from Pigeon Liver | | Descriptor: | CHLORIDE ION, MALIC ENZYME, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Zhang, H, Liang, T. | | Deposit date: | 2001-11-19 | | Release date: | 2002-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Pigeon Cytosolic Nadp+ -Dependent Malic Enzyme
Protein Sci., 11, 2002
|
|
1GZ4
 
 | | molecular mechanism of the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)(+)-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
8IMW
 
 | |
5HDT
 
 | | Human cohesin regulator Pds5B bound to a Wapl peptide | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Sister chromatid cohesion protein PDS5 homolog B, Wings apart-like protein homolog | | Authors: | Ouyang, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-09 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Structure of the human cohesin regulator Pds5 in complex with Wapl motif
To Be Published
|
|
7TLK
 
 | | Crystal Structure of K-Ras(G12S) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71102226 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
7TLG
 
 | | Crystal Structure of small molecule beta-lactone 5 covalently bound to K-Ras(G12S) | | Descriptor: | (3R,4R)-1-[7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(4S,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-4-yl]-3-hydroxypiperidine-4-carbaldehyde, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.80000722 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
3K6J
 
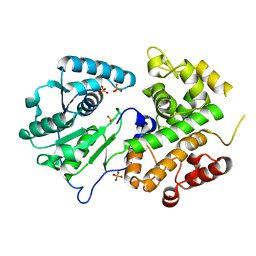 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
7TLE
 
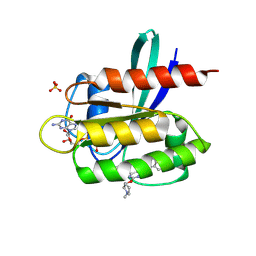 | | Crystal Structure of small molecule beta-lactone 1 covalently bound to K-Ras(G12S) | | Descriptor: | (3R,4R)-1-[7-(8-chloronaphthalen-1-yl)-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]-3-hydroxypiperidine-4-carbaldehyde, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98716748 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
7CCG
 
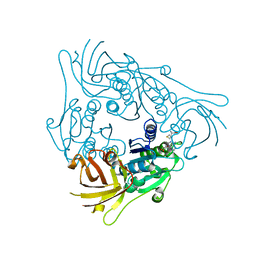 | |
7YKS
 
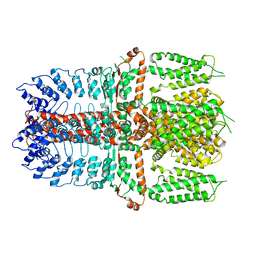 | |
7YKR
 
 | |
8W8Q
 
 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
