2RMP
 
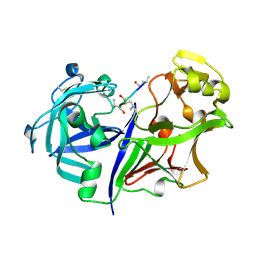 | | RMP-pepstatin A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MUCOROPEPSIN, PEPSTATIN, ... | | Authors: | Yang, J, Quail, J.W. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rhizomucor miehei aspartic proteinase complexed with the inhibitor pepstatin A at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5HM2
 
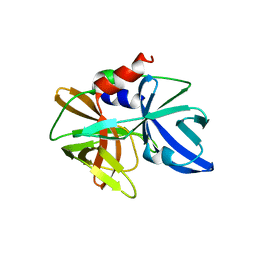 | |
7N1R
 
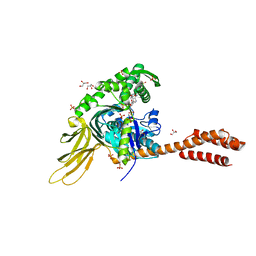 | | A novel and unique ATP hydrolysis to AMP by a human Hsp70 BiP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Yang, J, Musayev, F, Liu, Q. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel and unique ATP hydrolysis to AMP by a human Hsp70 Binding immunoglobin protein (BiP).
Protein Sci., 31, 2022
|
|
7YOC
 
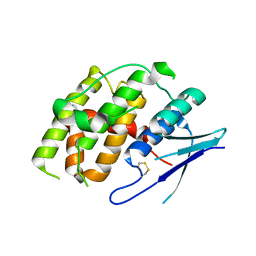 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YP0
 
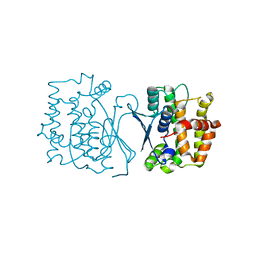 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
2WMO
 
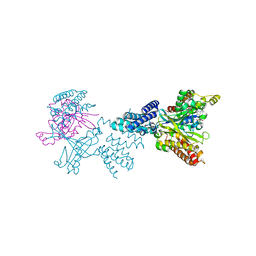 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WMN
 
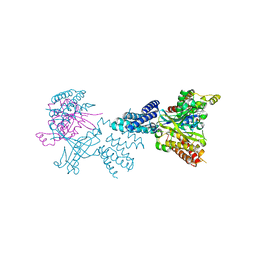 | | Structure of the complex between DOCK9 and Cdc42-GDP. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WM9
 
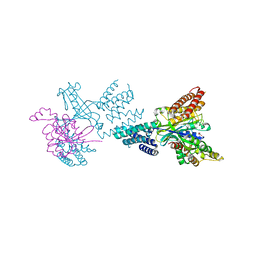 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
1H6G
 
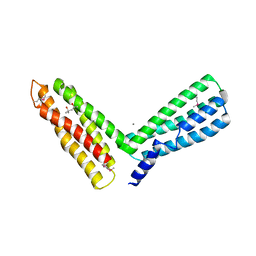 | | alpha-catenin M-domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-1 CATENIN, CALCIUM ION, ... | | Authors: | Yang, J, Dokurno, P, Tonks, N.K, Barford, D. | | Deposit date: | 2001-06-14 | | Release date: | 2001-08-07 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the M-Fragment of Alpha-Catenin: Implications for Modulation of Cell Adhesion.
Embo J., 20, 2001
|
|
6LPX
 
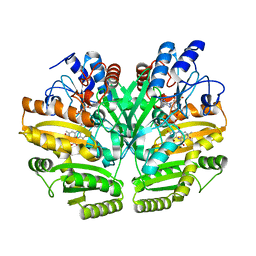 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with 2-oxoglutarate (2-OG) | | Descriptor: | 2-OXOGLUTARIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPP
 
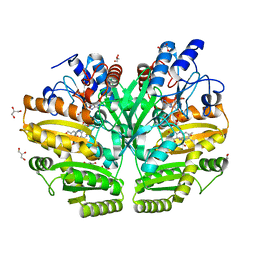 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-2-hydroxyglutarate (D-2-HG) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPU
 
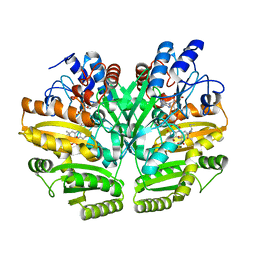 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPQ
 
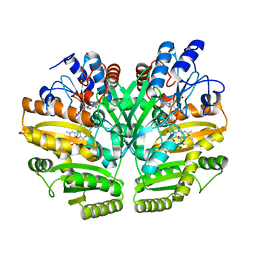 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-malate (D-MAL) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, D-MALATE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPN
 
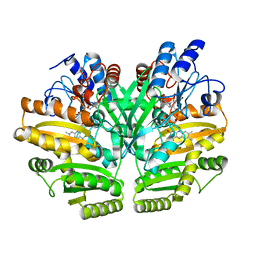 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in apo form | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
1F9S
 
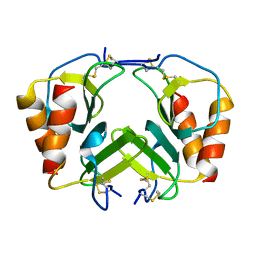 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 2 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1F9P
 
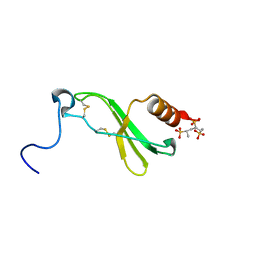 | | CRYSTAL STRUCTURE OF CONNECTIVE TISSUE ACTIVATING PEPTIDE-III(CTAP-III) COMPLEXED WITH POLYVINYLSULFONIC ACID | | Descriptor: | CONNECTIVE TISSUE ACTIVATING PEPTIDE-III, ETHANESULFONIC ACID | | Authors: | Yang, J, Faulk, T, Aster, R, Visentin, G, Edwards, B, Castor, C. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the CXC Chemokine, Connective Tissue Activating Peptide-III, Complexed with the Heparin Analogue, Polyvinylsulfonic Acid
To be Published
|
|
1F9R
 
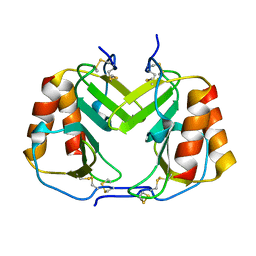 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 1 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1F9Q
 
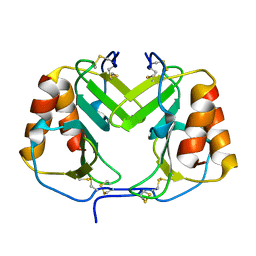 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-Type Reveals the Epitopes for the Heparin-Induced Thrombocytopenia Antibodies
To be Published
|
|
1FPR
 
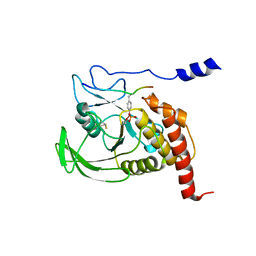 | | CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN THE CATALYTIC DOMAIN OF SHP-1 AND AN IN VITRO PEPTIDE SUBSTRATE PY469 DERIVED FROM SHPS-1. | | Descriptor: | PEPTIDE PY469, PROTEIN-TYROSINE PHOSPHATASE 1C | | Authors: | Yang, J, Cheng, Z, Niu, Z, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2000-08-31 | | Release date: | 2001-03-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 275, 2000
|
|
1GWZ
 
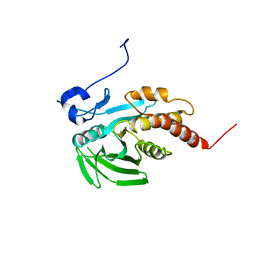 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
7D6N
 
 | |
7D6M
 
 | |
8IOM
 
 | |
4A2N
 
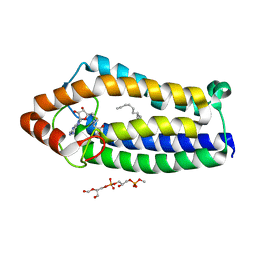 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
