6DZZ
 
 | | Cryo-EM Structure of the wild-type human serotonin transporter in complex with ibogaine and 15B8 Fab in the inward conformation | | Descriptor: | (5beta)-12-methoxyibogamine, 15B8 antibody heavy chain, 15B8 antibody light chain, ... | | Authors: | Yang, D, Coleman, J.A, Gouaux, E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569, 2019
|
|
2B1O
 
 | | Solution Structure of Ca2+-bound DdCAD-1 | | Descriptor: | CALCIUM ION, Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Sriskanthadevan, S, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7BZA
 
 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XCA
 
 | | Crystal structure of the porcine astrovirus capsid spike domain | | Descriptor: | CHLORIDE ION, Capsid protein spike domain, MAGNESIUM ION | | Authors: | Pan, L.X, Zhang, W.C, Yang, D.F, Yang, L.Y, Liu, H. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of porcine astrovirus capsid spike and spike/neutralizing antibody complexes.
To Be Published
|
|
3C90
 
 | | The 1.25 A Resolution Structure of Phosphoribosyl-ATP Pyrophosphohydrolase from Mycobacterium tuberculosis, crystal form II | | Descriptor: | Phosphoribosyl-ATP pyrophosphatase | | Authors: | Javid-Majd, F, Yang, D, Ioerger, T.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The 1.25 A resolution structure of phosphoribosyl-ATP pyrophosphohydrolase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7N7E
 
 | |
7N7D
 
 | |
6M19
 
 | | Template lasso peptide C24 mutant W14F | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-02-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
8YW5
 
 | | Cryo-EM structure of the retatrutide-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
6LCI
 
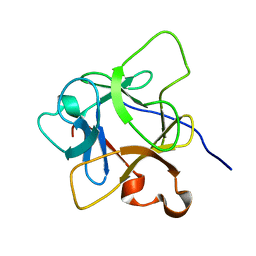 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
6LAG
 
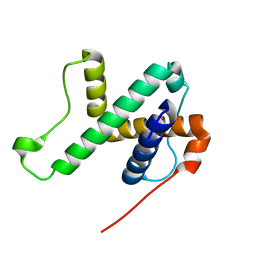 | | Solution structure of SPA-2 SHD | | Descriptor: | Spa2-like protein | | Authors: | Fan, J.S, Wong, J.Y, Zheng, P, Yang, D, Jedd, G. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spitzenkorper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds.
Nat Commun, 11, 2020
|
|
6L7K
 
 | | solution structure of hIFABP V60C/Y70C variant. | | Descriptor: | Fatty acid-binding protein, intestinal | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.
Biophys.J., 118, 2020
|
|
5WVP
 
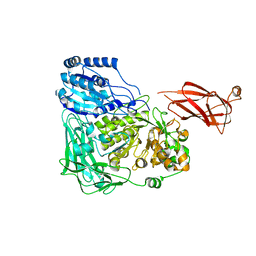 | | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii | | Descriptor: | Beta-glucosidase, beta-D-mannopyranose | | Authors: | Jiang, Z, Wu, S, Yang, D, Qin, Z, You, X, Huang, P. | | Deposit date: | 2016-12-28 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii
To Be Published
|
|
7O1E
 
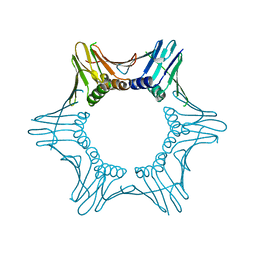 | | Crystal structure of PCNA from Chaetomium thermophilum | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Alphey, M.A, MacNeill, S, Yang, D. | | Deposit date: | 2021-03-29 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Non-canonical binding of the Chaetomium thermophilum PolD4 N-terminal PIP motif to PCNA involves Q-pocket and compact 2-fork plug interactions but no 3 10 helix.
Febs J., 290, 2023
|
|
6LMR
 
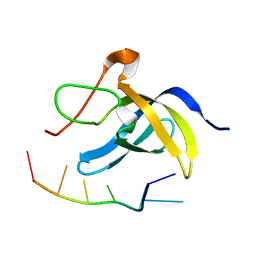 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
7MSV
 
 | |
7O1F
 
 | |
